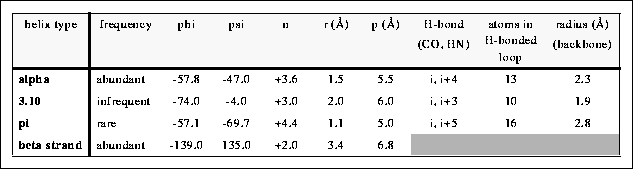
n - the number of residues per helical turn
r - the rise per helical residue
Q1. You can call a alpha-helix as
3.613 helix. What can you call a pi-helix in this
kind of nomenclature?
What are the n and r parameters for pi-helices?
Ans:
n - the number of residues per helical turn
r - the rise per helical residue
4.416 Helix
n-4.4
r-1.1 Angstroms
Q.2
Ans:
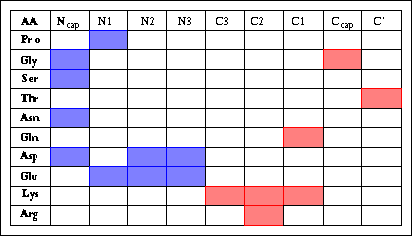
Helix boundary residues (the first and last helical residues)
are called Ncap and Ccap at the N- and C-terminus, respectively (Richardson
& Richardson, 1988). The proposed nomenclature uses the following definition:
...-N''-N'-Ncap-N1-N2-N3-............-C3-C2-C1-Ccap-C'-C''-...
But some amino acids have strong preferences, like this table shows.
Pro: For it's structure, I think the cis
conformation plays an important role! And it has a hydrogen donor in -NH.
Gly: It is flexible,
so it can be N-cap and C-cap.
Ser: The -NH in
Ncap will form a hydrogen bond to residue bellow.
Thr: The R group
has -OH, and the hydroxyl group can be a hydrogen bond aceptor.
Asn: Maybe the -NH
in R group arm can be a hydrogen donor in ncap position!
Gln: Like r group
of Asn, but Gln contribute it's C=O on R group arm to form the H-bond.
Asp & Glu: Appears
in many places in N-terminal, for it has COOH in R group.Can form new H-bond
with previous a.a.'s -NH .
Lys & Arg:The
-NH on R group arm can be a hydrogen donor for synthsis Hydrogen-Bond.
Q3. How does a prolin
residue lead to the kink of alpha-helices?
Ans: Pro (and hydroxyproline)
acts as helix breaker(or helix terminator) due to their unique structure,
with fixes the value of the Calpha-N-C bond angle.
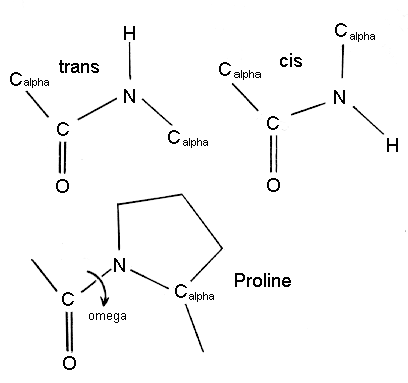
Proline kinked helices are almost exclusively long helices (>4
turns) which might be a requirement to overcome the effect of the kink.
Somewhat surprisingly, only one hydrogen bond (amide of the following residue,
i+1, to the carbonyl oxygen of i-3) was found to be broken due to the kink
in the helix.
Q4.
A
AEAAAKEAAAKEAAAKA
B AKAAAEKAAAEKAAAEA
C AEAAKAEAAKAEAAKA Blue: positively charged residues
D
AKAAEAKAAEAKAAEA
red: negatively
charged residues
were compared (S. Marqusee and R. L. Baldwin, Proc. Natl. Acad.
Sci. USA 84, 8898-8902,
1987). The a -amino and a-carboxyl groups
were blocked with acetyl and amide groups,
respectively. How would the first and second
peptides of each pair be expected to differ in their
helicity? What's the reasons for these differences?
Ans: A & B are i+4 helix, and C &
D are i+3 helix. The helicity of i+4 is better than i+3! So AB are better
than CD! According to the macrodipole, positive charges of N-terminal
is more stable. So A is better than B, and C is better than D.
Q5. Ion pairs in proteins involving
Arg residues have been observed to be energetically stronger
than those involving Lys residues. If this
were the case, in what ways might Arg and Lys residues
be used differently in proteins? How could
this hypothesis be tested? (Hint: D. B. Wigley et al.,
Biochem. Biophys. Res. Commun. 149,
927-929, 1987)
Ans:Arg forms a tighter ion pair with
a carboxylate group than dose Lys and is always for ion-pairs which are
not broken during turnover in naturally-occuring enzymes. The ion pair
with Arg was 20kJ/mol strong than the corresponding Lys-carboxylate
ion pair, so the bond of Lys is weak. From the six proteins in the paper(Table
1). The bond between the protein and carboxylate is always with Arg when
the protein carboxylate bond is not transformed during the
enzyme reaction.
The enzyme involved Lys residues in this case, I think it may be a
"transcarboxylase". The carboxylate is transformes during the
enzyme reaction. The distinction can be well illustration with
citrate synthase. Citrate contains three carbonxylates.
Two of these remain attach to the protein durind catalysis and make bonds
with Arg. The third, which is drived from the thioester in Acetyl-CoA is
bound to the enzyme by catalytic His.
Q6.a) This is a schematically
drawing of a four-stranded beta sheet. Please indicate the antiparallel
and parallel strands.
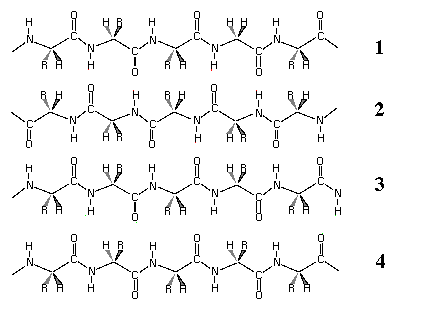
Ans:
1 ------->
2 <-------
3 ------->
4 ------->
antiparallel strans: 1-2, 2-3
parallel strans: 3-4
b) What is the diople moment of
a beta-sheet? Is it plausible that parallel and antiparallel sheets
could have substantially different dipole
interactions? (Hint: W. G. Hol et al., Nature 294,
532-536, 1981; P. T. van Duijnen et al., Biopolymers
24, 735-745, 1985)
Ans:In studying the diople moment of a
beta-sheet phenomenon electrostatically,we examined the arrangement of
peptide units in parallel beta-strands. From schematic drawings of hydrogen
bondind patterns in parallel beta-sheet(Fig 2 on the paper), it is obvious
that the N->H as well as the C->O diploes point backward with respect to
the N to C-terminal direction of the strands. The suggests that the parallel
beta-sheet has a significant overrall dipole moment, with the N-terminal
end of the strands corresponding to the positive ends of the dipole(usually
near C-terminal end).
Electrostatically, an energy difference between two kinds of
pleated sheet may be related to the fact that in an antiparallel sheet
all peptide dipole moments seem to cancel each other out; there is no residual
moment in either direction. Difference in electrostatic energy of ~0.8
kcal/mol between a parallel and antiparallel arrangement of three strands.
And "Yes", it is significant ! they have substantically different dipole
interactions.