Cardiotoxin III - Chinese cobra
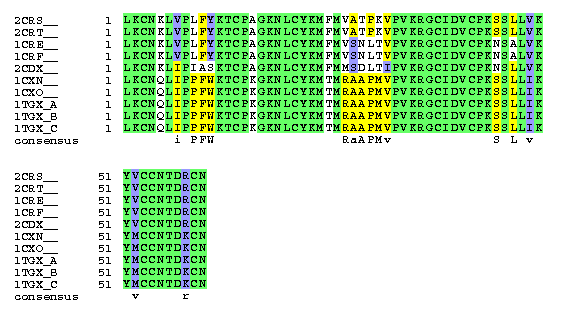
Proteins Sharing Sequence Homology with Cardiotoxin III
PDB:2crs__ - CARDIOTOXIN III (NMR, 13 STRUCTURES)
PDB:2crt__ - CARDIOTOXIN III (NMR, MINIMIZED AVERAGE STRUCTURE)
PDB:1cre__ - CARDIOTOXIN II (NMR, MINIMIZED AVERAGE STRUCTURE)
PDB:1crf__ - CARDIOTOXIN II (NMR, 12 STRUCTURES)
PDB:2cdx__ - CARDIOTOXIN CTX I (NMR, 11 STRUCTURES)
PDB:1cxn__ - CARDIOTOXIN GAMMA (NMR, MINIMIZED AVERAGE STRUCTURE)
PDB:1cxo__ - CARDIOTOXIN GAMMA (NMR, 9 STRUCTURES)
PDB:1tgx_A - TOXIN GAMMA (CARDIOTOXIN)
PDB:1tgx_B - TOXIN GAMMA (CARDIOTOXIN)
PDB:1tgx_C - TOXIN GAMMA (CARDIOTOXIN)
Secondary Structure
Chou-Fasman plot
60 aa;
. . . . . .
LKCNKLVPLFYKTCPAGKNLCYKMFMVATPKVPVKRGCIDVCPKSSLLVKYVCCNTDRCN
helix <---------> <-------------> <-------->
sheet EEEEEEEEE EEEEEEEEEE EEEE EEEEEEEEE
turns T T T TT
Residue totals: H: 36 E: 32 T: 5
percent: H: 60.0 E: 53.3 T: 8.3
Charge Distribution
1 0+00+00000 0+00000+00 00+0000000 +000++000- 000+00000+ 000000-+00
A. CHARGE CLUSTERS.
Positive charge clusters (cmin = 13/30 or 18/45 or 22/60): none
Negative charge clusters: not evaluated (frequency of - < 5%, too low)
Mixed charge clusters (cmin = 15/30 or 20/45 or 25/60): none
B. HIGH SCORING (UN)CHARGED SEGMENTS.
______________________________________
High scoring positive charge segments:
score= 2.00 frequency= 0.183 ( KR )
score= 0.00 frequency= 0.000 ( BZX )
score= -1.00 frequency= 0.783 ( LAGSVTIPNFQYHMCW )
score= -2.00 frequency= 0.033 ( ED )
Expected score/letter: -0.483
- now scoring for positive charge segments; Average information/letter: 0.430
Minimal length of displayed segments set to: 20
M_0.01= 13.07 (cv= 7.77, lambda= 0.52686, k= 0.16371, x= 5.30;
90% confidence interval for segment length: 23 +- 25)
M_0.05= 9.97 (x= 2.20)
# of segments (>=20 residues) exceeding M_0.05: none
______________________________________
High scoring negative charge segments:
score= 2.00 frequency= 0.033 ( ED )
score= 0.00 frequency= 0.000 ( BZX )
score= -1.00 frequency= 0.783 ( LAGSVTIPNFQYHMCW )
score= -2.00 frequency= 0.183 ( KR )
Expected score/letter: -1.083
- now scoring for negative charge segments; Average information/letter: 3.490
Minimal length of displayed segments set to: 20
M_0.01= 4.94 (cv= 2.54, lambda= 1.61122, k= 0.47671, x= 2.40;
90% confidence interval for segment length: 3 +- 3)
M_0.05= 3.92 (x= 1.38)
# of segments (>=20 residues) exceeding M_0.05: none
___________________________________
High scoring mixed charge segments:
score= 1.00 frequency= 0.217 ( KEDR )
score= 0.00 frequency= 0.000 ( BZX )
score= -1.00 frequency= 0.783 ( LAGSVTIPNFQYHMCW )
Expected score/letter: -0.567
- now scoring for mixed charge segments; Average information/letter: 1.051
Minimal length of displayed segments set to: 20
M_0.01= 6.07 (cv= 3.19, lambda= 1.28520, k= 0.40993, x= 2.89;
90% confidence interval for segment length: 11 +- 9)
M_0.05= 4.80 (x= 1.62)
# of segments (>=20 residues) exceeding M_0.05: none
________________________________
High scoring uncharged segments:
score= 1.00 frequency= 0.783 ( LAGSVTIPNFQYHMCW )
score= 0.00 frequency= 0.000 ( BZX )
score= -8.00 frequency= 0.217 ( KEDR )
Expected score/letter: -0.950
- now scoring for uncharged segments; Average information/letter: 0.173
Minimal length of displayed segments set to: 20
M_0.01= 32.53 (cv= 20.56, lambda= 0.19916, k= 0.10900, x= 11.97;
90% confidence interval for segment length: 54 +- 44)
M_0.05= 24.34 (x= 3.79)
# of segments (>=20 residues) exceeding M_0.05: none
C. CHARGE RUNS AND PATTERNS.
pattern (+)| (-)| (*)| (0)| (+0)| (-0)| (*0)|(+00)|(-00)|(*00)|
lmin0 5 | 3 | 6 | 29 | 10 | 6 | 10 | 11 | 6 | 12 |
lmin1 6 | 4 | 7 | 36 | 12 | 7 | 12 | 14 | 8 | 14 |
lmin2 7 | 4 | 8 | 39 | 13 | 8 | 14 | 15 | 9 | 16 |
There are no charge runs or patterns exceeding the given minimal lengths.
Run count statistics:
+ runs >= 3: 0
- runs >= 3: 0
* runs >= 4: 0
0 runs >= 20: 0




