 3. Its secondary structure predicted by Garnier method. Osgusthorpe, and Robinson, J. Mol. Biol.,(1978) 120:97-120
3. Its secondary structure predicted by Garnier method. Osgusthorpe, and Robinson, J. Mol. Biol.,(1978) 120:97-120
1. This is a known protein, and here are some of its descriptions:
HEADER Cardiotoxin 12-MAR-94 2CRS 2CRS 2
COMPND Cardiotoxin III (NMR, 13 STRUCTURES) 2CRS 3
SOURCE Taiwan COBRA (NAJA NAJA ATRA) Venom 2CRS 4
EXPDTA NMR 2CRS 5
AUTHOR R.BHASKARAN,C.C.HUANG,K.D.CHANG,C.YU 2CRS 6
REVDAT 1 01-Nov-94 2CRS 0 2CRS 7
JRNL
AUTH R.BHASKARAN,C.C.HUANG,K.D.CHANG,C.YU
SWISS-PROT ANNOTATION:
ID 11557
DE 11557, 60 bases.
number of residues: 60; molecular weight: 6.7 kdal
1 LKCNKLVPLF YKTCPAGKNL CYKMFMVATP KVPVKRGCID VCPKSSLLVK YVCCNTDRCN
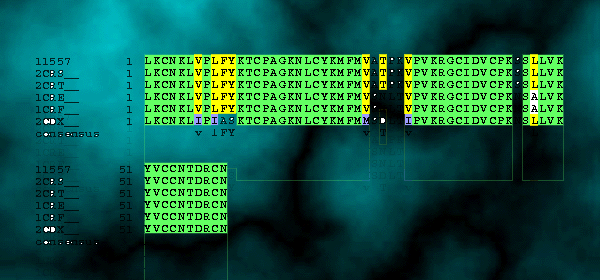
2. Multiple sequence Alignment for proteins that share homology with Cardiotoxin III
 3. Its secondary structure predicted by Garnier method. Osgusthorpe, and Robinson, J. Mol. Biol.,(1978) 120:97-120
3. Its secondary structure predicted by Garnier method. Osgusthorpe, and Robinson, J. Mol. Biol.,(1978) 120:97-120
4. CHARGE DISTRIBUTIONAL ANALYSIS
1 0+00+00000 0+00000+00 00+0000000 +000++000- 000+00000+ 000000-+00
Positive charge clusters (cmin = 13/30 or 18/45 or 22/60): none
Negative charge clusters: not evaluated (frequency of - < 5%, too low)
Mixed charge clusters (cmin = 15/30 or 20/45 or 25/60): none
High scoring positive charge segments:
score= 2.00 frequency= 0.183 ( KR )
score= 0.00 frequency= 0.000 ( BZX )
score= -1.00 frequency= 0.783 ( LAGSVTIPNFQYHMCW )
score= -2.00 frequency= 0.033 ( ED )
Minimal length of displayed segments set to: 20
M_0.01= 13.07 (cv= 7.77, lambda= 0.52686, k= 0.16371, x= 5.30;90% confidence interval for segment length: 23 +- 25)
M_0.05= 9.97 (x= 2.20)
# of segments (>=20 residues) exceeding M_0.05: none
score= 2.00 frequency= 0.033 ( ED )
score= 0.00 frequency= 0.000 ( BZX )
score= -1.00 frequency= 0.783 ( LAGSVTIPNFQYHMCW )
score= -2.00 frequency= 0.183 ( KR )
Minimal length of displayed segments set to: 20
M_0.01= 4.94 (cv= 2.54, lambda= 1.61122, k= 0.47671, x= 2.40;90% confidence interval for segment length: 3 +- 3)
M_0.05= 3.92 (x= 1.38)
# of segments (>=20 residues) exceeding M_0.05: none
score= 1.00 frequency= 0.217 ( KEDR )
score= 0.00 frequency= 0.000 ( BZX )
score= -1.00 frequency= 0.783 ( LAGSVTIPNFQYHMCW )
Minimal length of displayed segments set to: 20
M_0.01= 6.07 (cv= 3.19, lambda= 1.28520, k= 0.40993, x= 2.89;90% confidence interval for segment length: 11 +- 9)
M_0.05= 4.80 (x= 1.62)
# of segments (>=20 residues) exceeding M_0.05: none
score= 1.00 frequency= 0.783 ( LAGSVTIPNFQYHMCW )
score= 0.00 frequency= 0.000 ( BZX )
score= -8.00 frequency= 0.217 ( KEDR )
M_0.01= 32.53 (cv= 20.56, lambda= 0.19916, k= 0.10900, x= 11.97; 90% confidence interval for segment length: 54 +- 44)
M_0.05= 24.34 (x= 3.79)
# of segments (>=20 residues) exceeding M_0.05: none
pattern (+)| (-)| (*)| (0)| (+0)| (-0)| (*0)|(+00)|(-00)|(*00)|
lmin0 5 | 3 | 6 | 29 | 10 | 6 | 10 | 11 | 6 | 12 |
lmin1 6 | 4 | 7 | 36 | 12 | 7 | 12 | 14 | 8 | 14 |
lmin2 7 | 4 | 8 | 39 | 13 | 8 | 14 | 15 | 9 | 16 |
There are no charge runs or patterns exceeding the given minimal lengths.
Run count statistics:
+ runs >= 3: 0
- runs >= 3: 0
0 runs >= 20: 0