hooomework5
HOMEWORK5

1.The accession number of mouse
LEF-1 and TCF-1 in SWISS-PROT
LEF-1:P27782......
TCF-1:Q00417......
2.The name and the residue
number of the same DNA-binding domain corresponding
to the full-length protein
Name:HMG BOX
LEF-1:297-365......
TCF-1:188-256......
3.The AA seequence of this
DNA-binding domain of each protein
LEF-1:
the DNA-binding domain is colored by green
MPQLSGGGGG GDPELCATDE MIPFKDEGDP QKEKIFAEIS HPEEEGDLAD IKSSLVNESE
IIPASNGHEV VRQAPSSQEP YHDKAREHPD EGKHPDGGLY NKGPSYSSYS GYIMMPNMNS
DPYMSNGSLS PPIPRTSNKV PVVQPSHAVH PLTPLITYSD EHFSPGSHPS HIPSDVNSKQ
GMSRHPPAPE IPTFYPLSPG GVGQITPPIG WQGQPVYPIT GGFRQPYPSS LSGDTSMSRF
SHHMIPGPPG PHTTGIPHPA IVTPQVKQEH PHTDSDLMHV KPQHEQRKEQ EPKRPHIKKP
LNAFMLYMKE MRANVVAECT LKESAAINQI LGRRWHALSR EEQAKYYELA RKERQLHMQL
YPGWSARDNY GKKKKRKREK LQESTSGTGP RMTAAYI.........................
TCF-1:the DNA-binding domain is colored by blue
MYKETVYSAF NLLMPYPPAS GAGQHPQPQP PLHNKPGQPP HGVPQLSPLY EHFSSPHPTP
APADISQKQG VHRPLQTPDL SGFYSLTSGS MGQLPHTVSW PSPPLYPLSP SCGYRQHFPA
PTAAPGAPYP RFTHPSLMLG SGVPGHPAAI PHPAIVPSSG KQELQPYDRN LKTQAEPKAE
KEAKKPVIKK PLNAFMLYMK EMRAKVIAEC TLKESAAINQ ILGRRWHALS REEQAKYYEL
ARKERQLHMQ LYPGWSARDN YGKKKRRSRE KHQESTTGGK RNAFGTYPEK AAAPAPFLPMTVL
4.The pI/Mw and the predicted secondary of this DNA-binding domain of LEF-1(from ProtParam)
Molecular weight:8239.6
Theoretical pI:9.66
The predicted secondary is alpha-helix
5.The hydropathicity of this DNA-binding domain of TCF-1(from ProtScale)
Using the scale Hphob. / Kyte & Doolittle, the individual values for the 20 amino acids are:
Ala: 1.800 Arg: -4.500 Asn: -3.500 Asp: -3.500 Cys: 2.500 Gln: -3.500
Glu: -3.500 Gly: -0.400 His: -3.200 Ile: 4.500 Leu: 3.800 Lys: -3.900
Met: 1.900 Phe: 2.800 Pro: -1.600 Ser: -0.800 Thr: -0.700 Trp: -0.900
Tyr: -1.300 Val: 4.200 Asx: -3.500 Glx: -3.500 Xaa: -0.490 ...........
Weights for window positions 1,..,9, using linear weight variation model:
1 2 3 4 5 6 7 8 9
1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00
edge center edge
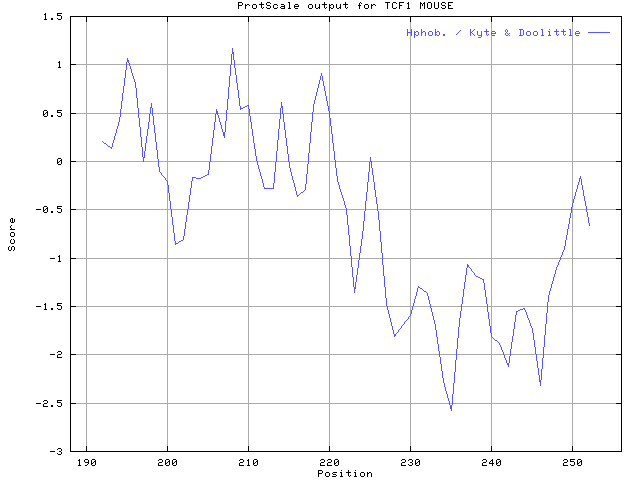
6.The chymotrypsin cleavage of TCF-1(from PeptideMass)
You have selected TCF1_MOUSE from SWISS-PROT:
T-CELL-SPECIFIC TRANSCRIPTION FACTOR 1 (TCF-1) (T-CELL FACTOR 1)
(TRANSCRIPTION FACTOR-7).
[Theoretical pI: 9.79 / Mw (average mass): 33608.54 / Mw (monoisotopic mass): 33587.19]
| mass | position |
#MC | peptide sequence |
| 3987.117 |
262-297 |
1 |
GKKKRRSREKHQESTTGGKR NAFGTYPEKAAAPAPF |
| 3456.738 |
14-46 |
1 |
MPYPPASGAGQHPQPQPPLH NKPGQPPHGVPQL |
| 3392.775 |
256-284 |
1 |
SARDNYGKKKRRSREKHQES TTGGKRNAF |
| 2874.486 |
54-80 |
1 |
SSPHPTPAPADISQKQGVHR PLQTPDL |
| 2859.678 |
168-192 |
1 |
DRNLKTQAEPKAEKEAKKPV IKKPL |
| 2791.453 |
140-167 |
1 |
GSGVPGHPAAIPHPAIVPSS GKQELQPY |
| 2733.386 |
51-75 |
1 |
EHFSSPHPTPAPADISQKQG VHRPL |
| 2693.571 |
172-195 |
1 |
KTQAEPKAEKEAKKPVIKKP LNAF |
| 2686.472 |
262-284 |
0 |
GKKKRRSREKHQESTTGGKR NAF |
| 2575.230 |
109-132 |
1 |
SPSCGYRQHFPAPTAAPGAP YPRF |
| 2516.362 |
139-164 |
1 |
LGSGVPGHPAAIPHPAIVPS SGKQEL |
| 2516.295 |
115-137 |
1 |
RQHFPAPTAAPGAPYPRFTH PSL |
| 2403.278 |
140-164 |
0 |
GSGVPGHPAAIPHPAIVPSS GKQEL |
| 2361.423 |
172-192 |
0 |
KTQAEPKAEKEAKKPVIKKP L |
| 2320.216 |
54-75 |
0 |
SSPHPTPAPADISQKQGVHR PL |
| 2171.222 |
202-222 |
1 |
RAKVIAECTLKESAAINQIL |
| 2083.043 |
13-32 |
1 |
LMPYPPASGAGQHPQPQPPL |
| 1981.019 |
115-132 |
0 |
RQHFPAPTAAPGAPYPRF |
| 1978.969 |
87-105 |
1 |
TSGSMGQLPHTVSWPSPPL |
| 1969.959 |
14-32 |
0 |
MPYPPASGAGQHPQPQPPL |
| 1888.996 |
92-108 |
1 |
GQLPHTVSWPSPPLYPL |
| 1802.966 |
33-49 |
1 |
HNKPGQPPHGVPQLSPL |
| 1660.840 |
285-300 |
1 |
GTYPEKAAAPAPFLPM |
| 1641.918 |
213-226 |
1 |
KESAAINQILGRRW |
| 1515.795 |
92-105 |
0 |
GQLPHTVSWPSPPL |
| 1505.797 |
33-46 |
0 |
HNKPGQPPHGVPQL |
| 1491.802 |
200-212 |
1 |
KEMRAKVIAECTL |
| 1331.670 |
227-237 |
1 |
HALSREEQAKY |
| 1319.663 |
285-297 |
0 |
GTYPEKAAAPAPF |
| 1228.538 |
252-261 |
1 |
YPGWSARDNY |
| 1173.553 |
230-238 |
1 |
SREEQAKYY |
| 1168.637 |
241-249 |
1 |
ARKERQLHM |
| 1142.664 |
239-247 |
1 |
ELARKERQL |
| 1103.624 |
203-212 |
0 |
RAKVIAECTL |
| 1086.615 |
213-222 |
0 |
KESAAINQIL |
| 1010.490 |
230-237 |
0 |
SREEQAKY |
7.The Prosite scanning of LEF-1
[1] PDOC00001 PS00001 ASN_GLYCOSYLATION
N-glycosylation site
Number of matches: 2
1 57-60 NESE
2 126-129 NGSL
[2] PDOC00005 PS00005 PKC_PHOSPHO_SITE
Protein kinase C phosphorylation site
Number of matches: 3
1 137-139 SNK
2 320-322 TLK
3 365-367 SAR
[3] PDOC00006 PS00006 CK2_PHOSPHO_SITE
Casein kinase II phosphorylation site
Number of matches: 7
1 40-43 SHPE
2 76-79 SSQE
3 157-160 TYSD
4 273-276 TDSD
5 320-323 TLKE
6 339-342 SREE
7 365-368 SARD
[4] PDOC00007 PS00007 TYR_PHOSPHO_SITE
Tyrosine kinase phosphorylation site
93-100 KHPDGGLY
[5] PDOC00008 PS00008 MYRISTYL
N-myristoylation site
Number of matches: 4
1 6-11 GGGGGG
2 7-12 GGGGGD
3 97-102 GGLYNK
4 166-171 GSHPSH
[6] PDOC00009 PS00009 AMIDATION
Amidation site
Number of matches: 2
1 331-334 LGRR
2 370-373 YGKK
 ¦^º¶
....................................
.........................................................................
........... NEXT HOMEWORK
¦^º¶
....................................
.........................................................................
........... NEXT HOMEWORK


 ¦^º¶
....................................
.........................................................................
...........
¦^º¶
....................................
.........................................................................
...........