1.
Find the structure of prion from PDB.
- List the following information : (1)ID number, (2)Authors, (3)The number
of residules in one monomer, (4)The disulfide bonds.
- Put the structure of prion on your homepage. Show different color for
different secondary stucture in cartoon form with sidechains on (helix in
blue, sheets in red). Show the script that makes the final picture.
|
|
- ID No.=1AG2
Authors=M.Billeter, R.Riek, G.Winder, K.Muthrich, S.Hornemann,
R.Glockshuber
No. of Residules=103
No. of Disulfide Bond=1
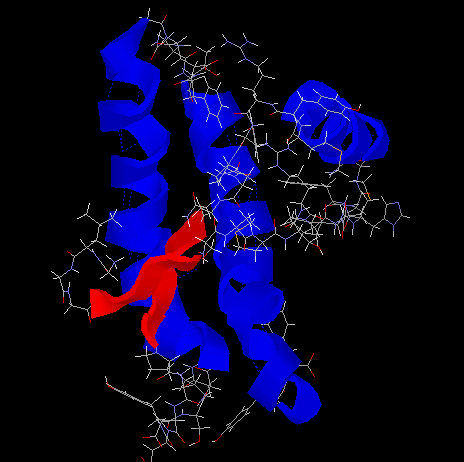
- The structure of Prion from PDB (edited with RasMol):
The Script:
RasMol$ select helix
RasMol$ wireframe off (Display Cartoon)
RasMol$ color red
RasMol$ select sheet
RasMol$ wireframe off (Display Cartoon)
RasMol$ color blue
2.
Trascription of the
ant gene during lytic growth of bacteriophage P22 is regulated by the
the cooperative binding of two Arc repressor dimers to a 21-base-pair operator
site. Please find the co-crystal stucture of this Arc tetramer-operator
complex from PDB.
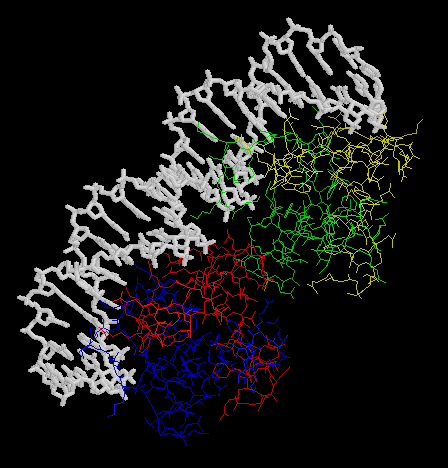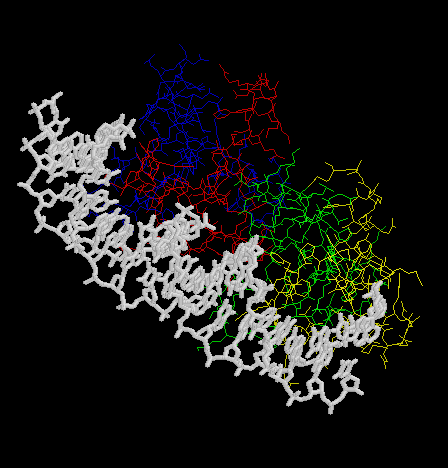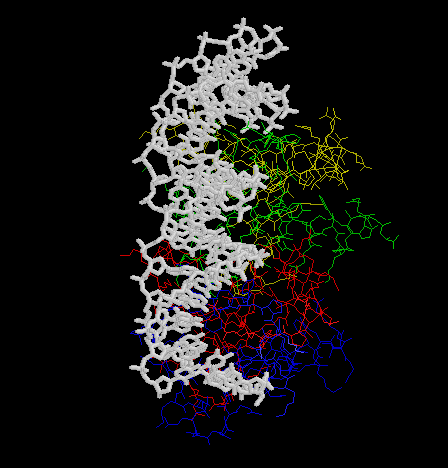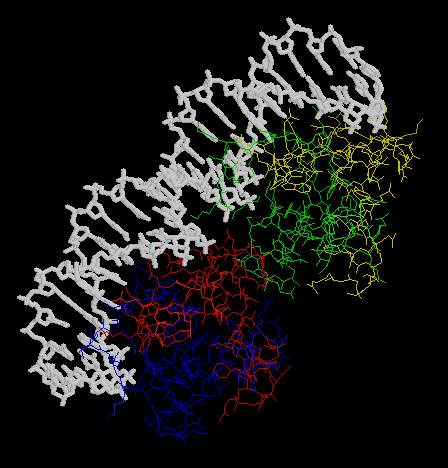
- Make an animation (by the Gifbuilder) of whole complex.
- Show the DNA sequence of the Arc operator.
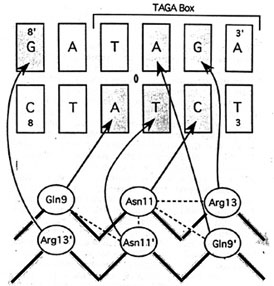
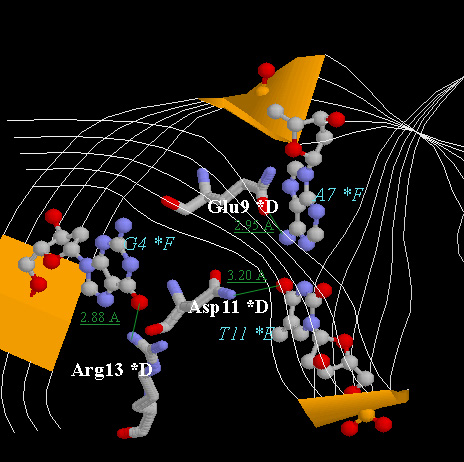
- The picture below is the hydrogen bonds between beta-sheet side chains
DNA bases. Show three side chains (gln9, asn11, arg13) from one monomer and
corresponding DNA bases of them on your home page. Color three side chain with
with different color. Measure the distance between possible hydrogen bonds.
|
|
- The animation (by Gifcon) of whole complex:
- The DNA sequenece of Arc operator:
*F-5'-A A T G A T A G A A G C A C T C T A C T A T- 3'
*E-3'-T T A C T A T C T T C G T G A G A T G A T A- 5'
-
| | |



