| Tools: | translating tools in ExPASy |
| Result: | choose the predicted sequence which contains the longest sequence between Met and Stop codon |
| 5`3` Frame1 A S T G L A S G L R S R P P P E A S S L T T Met D P R K V S E L R A F V K Met C R Q D P S V L H T E E Met R F L R E W V E S Met G G K V P P A T H K A K S E E N T K E E K R D K T T E D N I K T E E P S S E E S D L E I D N E G V I E A D T D A P Q E Met G D E N A E I T E A Met Met D E A N E K K G A A I D A L N D G E L Q K A I D L F T D A I K L N P R L A I L Y A K R A S V F V K L Q K P N A A I R D C D R A I E I N P D S A Q P Y K W R G K A H R L L G H W E E A A R D L A L A C K L D Y D E D A S A Met L R E V Q P R A Q K I A E H R R K Y E R K R E E R E I K E R I E R V K K A R E E H E K A Q R E E E A R R Q S G S Q F G S F P G G F P G G Met P G N F P G G Met P G Met G G A Met P G Met A G Met P G L N E I L S D P E V L A A Met Q D P E V Met V A F Q D V A Q N P S N Met S K Y Q N N P K V Met N L I S K L S A K F G G H S Stop C Q S P C Stop Met K N S L A H L L D V A I I Q T S V P L T S P E S W G A S K I I P T L C I I C G Stop G I L Q W F A I R V F I Q I Met F S Y Stop E L Q T Stop N I F Q P Stop T Y F L K I Stop G Met S I P T F F V T N L F G F F L L N Y W A R K V N V D D L L L S Stop Met K Stop R F V S G K Q I K H I Stop V D Stop V G H T V T A T S Stop I V F N V L L H N D L F Q Stop I F W D H Q K K K K K K K |
| Tools: | Direct WU-BLAST submission at EMBNet-CH (Lausanne, Switzerland) |
| Direct BLAST submission at NCBI (Bethesda, USA) | |
| Result: | This a HSC70-INTERACTING PROTEIN of Rattus norvegicus or Noway Rat. |
| Tools: | ProtParam |
| Results: | Total number of negatively charged residues (Asp + Glu): 69 |
| Total number of positively charged residues (Arg + Lys): 56 |
| Tools: | Hphob. / Eisenberg et al. in ProtScale |
| Results: | 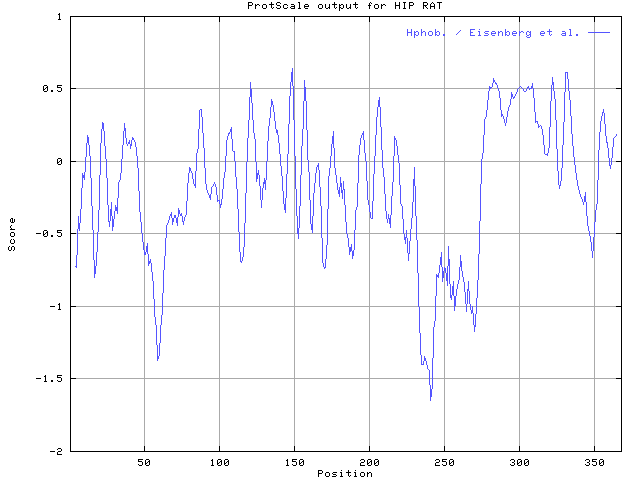
|
| Tools: | Prosite |
| Results: | There are five possible fonction foud |
[1] PDOC00001 PS00001 ASN_GLYCOSYLATION
N-glycosylation site
343-346 NMSK
[2] PDOC00004 PS00004 CAMP_PHOSPHO_SITE
cAMP- and cGMP-dependent protein kinase phosphorylation site
Number of matches: 3
1 4-7 RKVS
2 152-155 KRAS
3 270-273 RRQS
[3] PDOC00005 PS00005 PKC_PHOSPHO_SITE
Protein kinase C phosphorylation site
Number of matches: 2
1 46-48 THK
2 361-363 SAK
[4] PDOC00006 PS00006 CK2_PHOSPHO_SITE
Casein kinase II phosphorylation site
Number of matches: 5
1 55-58 TKEE
2 63-66 TTED
3 74-77 SSEE
4 78-81 SDLE
5 317-320 SDPE
[5] PDOC00008 PS00008 MYRISTYL
N-myristoylation site
Number of matches: 10
1 86-91 GVIEAD
2 274-279 GSQFGS
3 278-283 GSFPGG
4 282-287 GGFPGG
5 286-291 GGMPGN
6 287-292 GMPGNF
7 290-295 GNFPGG
8 294-299 GGMPGM
9 298-303 GMGGAM
10 301-306 GAMPGM
|
| Tools: | Protein Colourer |
| Results: | AGLPV FYW DENQRHSTK CM 
|