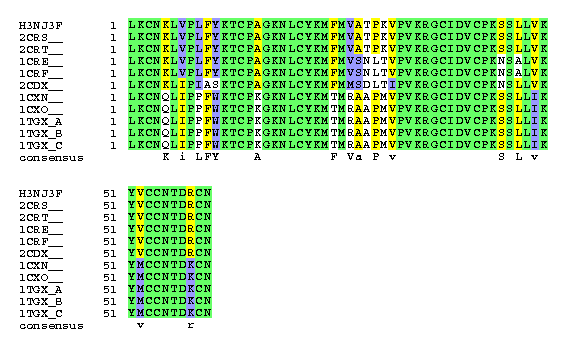
(1)
SO, it is not a new one. Three proteins in the protein detabase were found to be
indentical (100%) with these sequence.
search result
mktlllttvvvtivcldleytlkcnklvplfyktcpagknlcykmfmvatpkvpvkrgcidvcpkssllvkyvccntdrcn
Length = 81 Score = 340 (163.9 bits), Expect = 1.2e-44, P = 1.2e-44 Identities = 60/60
(100%), Positives = 60/60 (100%)
mktllltlvvvtivcldlgytlkcnklvplfyktcpagknlcykmfmvatpkvpvkrgcidvcpkssllvkyvccntdrcn
Length = 81 Score = 340 (163.9 bits), Expect = 1.2e-44, P = 1.2e-44 Identities = 60/60
(100%), Positives = 60/60 (100%)
lkcnklvplfyktcpagknlcykmfmvatpkvpvkrgcidvcpkssllvkyvccntdrcn
Length = 60 Score = 340 (163.9 bits), Expect = 1.6e-44, P = 1.6e-44 Identities = 60/60
(100%), Positives = 60/60 (100%)
. . . . . .
LKCNKLVPLFYKTCPAGKNLCYKMFMVATPKVPVKRGCIDVCPKSSLLVKYVCCNTDRCN
helix <---------> <-------------> <-------->
sheet EEEEEEEEE EEEEEEEEEE EEEE EEEEEEEEE
turns T T T TT
Residue totals: H: 36 E: 32 T: 5
percent: H: 60.0 E: 53.3 T: 8.3
. 10 . 20 . 30 . 40 . 50 . 60
LKCNKLVPLFYKTCPAGKNLCYKMFMVATPKVPVKRGCIDVCPKSSLLVKYVCCNTDRCN
helix H HHH HHHH
sheet EEEEEEEEE EEEEE E E EEEEE EEEEEEEE
turns TTT TTTT T T TT TTTT TTTTTT
coil C C
Residue totals: H: 8 E: 29 T: 21 C: 2
0 + 0 0 + 0 0 0 0 0 0 + 0 0 0 0 0 + 0 0 0 0 + 0 0 0 0 0 0 0 + 0 0 0 + + 0 0 0 - 0 0 0 + 0 0 0 0 0 + 0 0 0 0 0 0 - + 0 0 L K C N K L V P L F Y K T C P A G K N L C Y K M F M V A T P K V P V K R G C I D V C P K S S L L V K Y V C C N T D R C N