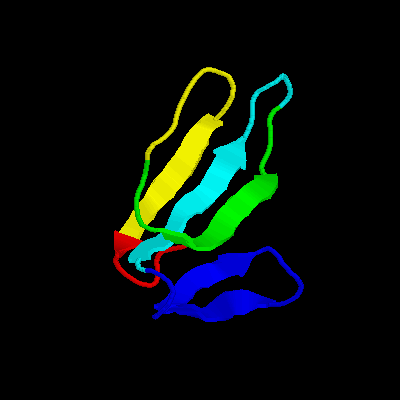
 2crt (minimized average structure)
2crt (minimized average structure)
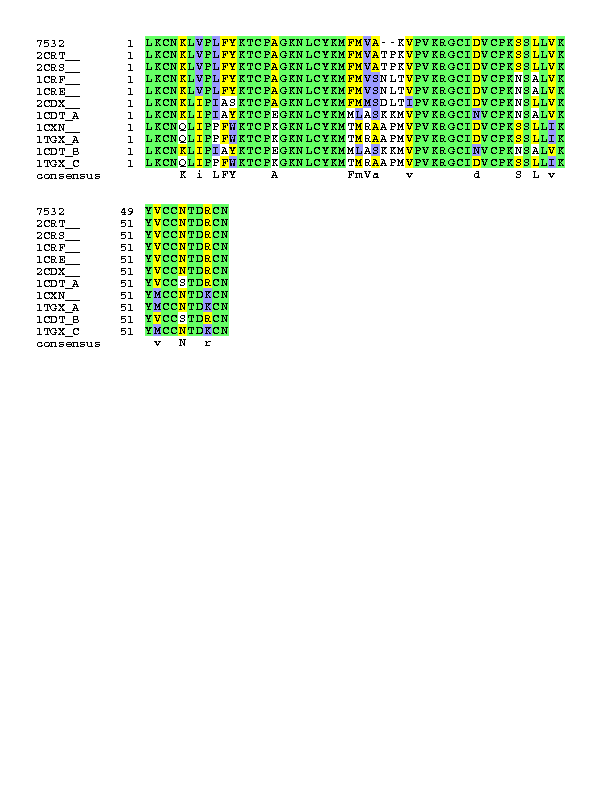
 (2) 含有此sequence之蛋白質如下:
(2) 含有此sequence之蛋白質如下:
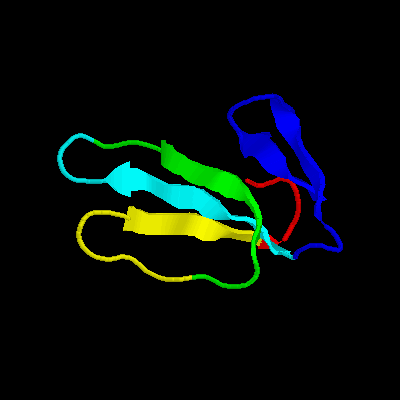
Cardiotoxin iii (NMR minimized average structure)
Cardiotoxin iii (NMR 13 structures)
Cardiotoxin ii (NMR 12 structures)
Cardiotoxin ii (NMR minimized average structure)
Cardiotoxin ctx i (NMR 11 structures)
Cardiotoxin v=4===II== (toxin III)
Cardiotoxin gamma (NMR minimized average structure)
Toxin gamma (cardiotoxin)
Cardiotoxin v=4===II== (toxin III)
Toxin gamma (cardiotoxin) LKCNKLVPLFYKTCPAGKNLCYKMFMVA--KVPVKRGCIDVCPKSSLLVKYVCCNTDRCN >2CRT__ LKCNKLVPLFYKTCPAGKNLCYKMFMVATPKVPVKRGCIDVCPKSSLLVKYVCCNTDRCN >2CRS__ LKCNKLVPLFYKTCPAGKNLCYKMFMVATPKVPVKRGCIDVCPKSSLLVKYVCCNTDRCN >1CRF__ LKCNKLVPLFYKTCPAGKNLCYKMFMVSNLTVPVKRGCIDVCPKNSALVKYVCCNTDRCN >1CRE__ LKCNKLVPLFYKTCPAGKNLCYKMFMVSNLTVPVKRGCIDVCPKNSALVKYVCCNTDRCN >2CDX__ LKCNKLIPIASKTCPAGKNLCYKMFMMSDLTIPVKRGCIDVCPKNSLLVKYVCCNTDRCN >1CDT_A LKCNKLIPIAYKTCPEGKNLCYKMMLASKKMVPVKRGCINVCPKNSALVKYVCCSTDRCN >1CXN__ LKCNQLIPPFWKTCPKGKNLCYKMTMRAAPMVPVKRGCIDVCPKSSLLIKYMCCNTDKCN >1TGX_A LKCNQLIPPFWKTCPKGKNLCYKMTMRAAPMVPVKRGCIDVCPKSSLLIKYMCCNTDKCN >1CDT_B LKCNKLIPIAYKTCPEGKNLCYKMMLASKKMVPVKRGCINVCPKNSALVKYVCCSTDRCN >1TGX_C LKCNQLIPPFWKTCPKGKNLCYKMTMRAAPMVPVKRGCIDVCPKSSLLIKYMCCNTDKCN
 (3) Their secondary structure (toxin and cardiotoxin iii) :
Garnier plot of toxin
58 aa; DCH = 0, DCS = 0
. 10 . 20 . 30 . 40 . 50
LKCNKLVPLFYKTCPAGKNLCYKMFMVAKVPVKRGCIDVCPKSSLLVKYVCCNTDRCN
helix H HHHHHHHHHHHHH
sheet EEEEEEEEE EEEEE EEEEEEEE
turns TTT TTTT TTTT TTTT TTTTTT
coil C
Residue totals: H: 14 E: 22 T: 21 C: 1
percent: H: 33.3 E: 52.4 T: 50.0 C: 2.4
Garnier plot of Cardiotoxin iii (NMR, minimized average structure)
60 aa; DCH = 0, DCS = 0
. 10 . 20 . 30 . 40 . 50 . 60
LKCNKLVPLFYKTCPAGKNLCYKMFMVATPKVPVKRGCIDVCPKSSLLVKYVCCNTDRCN
helix H HHH HHHH
sheet EEEEEEEEE EEEEE E E EEEEE EEEEEEEE
turns TTT TTTT T T TT TTTT TTTTTT
coil C C
(3) Their secondary structure (toxin and cardiotoxin iii) :
Garnier plot of toxin
58 aa; DCH = 0, DCS = 0
. 10 . 20 . 30 . 40 . 50
LKCNKLVPLFYKTCPAGKNLCYKMFMVAKVPVKRGCIDVCPKSSLLVKYVCCNTDRCN
helix H HHHHHHHHHHHHH
sheet EEEEEEEEE EEEEE EEEEEEEE
turns TTT TTTT TTTT TTTT TTTTTT
coil C
Residue totals: H: 14 E: 22 T: 21 C: 1
percent: H: 33.3 E: 52.4 T: 50.0 C: 2.4
Garnier plot of Cardiotoxin iii (NMR, minimized average structure)
60 aa; DCH = 0, DCS = 0
. 10 . 20 . 30 . 40 . 50 . 60
LKCNKLVPLFYKTCPAGKNLCYKMFMVATPKVPVKRGCIDVCPKSSLLVKYVCCNTDRCN
helix H HHH HHHH
sheet EEEEEEEEE EEEEE E E EEEEE EEEEEEEE
turns TTT TTTT T T TT TTTT TTTTTT
coil C C
 (4)
CHARGE DISTRIBUTIONAL ANALYSIS
1 0+00+00000 0+00000+00 00+0000000 +000++000- 000+00000+ 000000-+00
A. CHARGE CLUSTERS.
Positive charge clusters (cmin = 13/30 or 18/45 or 22/60): none
Negative charge clusters: not evaluated (frequency of - < 5%, too low)
Mixed charge clusters (cmin = 15/30 or 20/45 or 25/60): none
B. HIGH SCORING (UN)CHARGED SEGMENTS.
(4)
CHARGE DISTRIBUTIONAL ANALYSIS
1 0+00+00000 0+00000+00 00+0000000 +000++000- 000+00000+ 000000-+00
A. CHARGE CLUSTERS.
Positive charge clusters (cmin = 13/30 or 18/45 or 22/60): none
Negative charge clusters: not evaluated (frequency of - < 5%, too low)
Mixed charge clusters (cmin = 15/30 or 20/45 or 25/60): none
B. HIGH SCORING (UN)CHARGED SEGMENTS.
 回到上一頁
回到上一頁