It is not
a new protein.
The name
of this protein is Oryza sativa lipid transfer
protein.
Total number
of negatively
charged residues (Asp + Glu): 6
Total number
of positively
charged residues (Arg + Lys):12
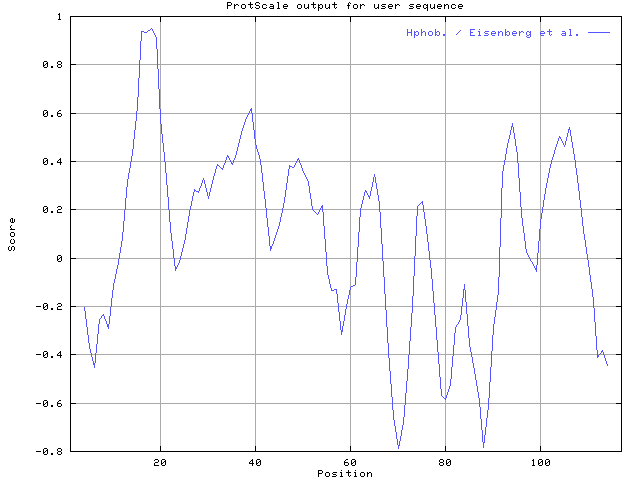
The hydrophobicity
map for this protein using Eisenberg hydrophobicity scale with window size
7. The relative weight of the window edges compared to the window center
should set to 40%.

There are hydrophobic regions in the protein. It may be a transmembrane region.
Possible functions
or pattern of this protein.
[1] PDOC00006
PS00006 CK2_PHOSPHO_SITE
Casein kinase
II phosphorylation site
66-69 TTAD
[2] PDOC00008
PS00008 MYRISTYL
N-myristoylation
site
Number of
matches: 3
1 29-34 GQVVST
2 46-51 GVAPTG
3 52-57 GCCDGV
[3] PDOC00516
PS00597 PLANT_LTP
Plant lipid
transfer proteins signature
95-116 IPSKCGVNIPYAISPSTDCSRV
Color the
protein by the hydrophobicity of the amino acids.
MARAGHNKYVARVMVVALLLAARAPVTCGQVVSTWAPCIMYADGEGVAPTGGCCDGVRTLNSAAATTADR
QTTCACLKQQTKAMGRLRPDHVAGIPSKCGVNIPYAISPSTDCSRVH
Total number
of ALIVMW in sequence : 45