











The answers of questions 1 are solved and reconstituted as the table Questions 1 a.) PDB ID number. b.) Name of the authors. c.) Expression system used by authors. d.) Residue number. e.) Number of helix. f.) The starting residue and ending residue of each helix. g.) Number of b-sheet.
| 1EMA | Fluorescent Protein | date | Aug 01, 1996 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| title | Green Fluorescent Protein From Aequorea Victoria | ||||||||||||||
| authors | M.Ormo, A.B.Cubitt, K.Kallio, L.A.Gross, R.Y.Tsien, S.J.Remington | ||||||||||||||
| compound | source | ||||||||||||||
| Molecule: Green Fluorescent
Protein
Chain: Null Engineered: Yes Mutation: 65 - 67 Replaced By Cro, Q80r Other_details: The Protein Contains Six Se-Methionines. Of These, The N-Terminal Met And Met 233 Are Not Present In The Entry |
Organism_scientific: Aequorea
Victoria
Organism_common: Jellyfish Plasmid: Prsetb (Invitrogen) Expression_system: Escherichia Coli Expression_system_strain: Jm109 (De3) Other_details: The N-Terminal His-Tag Has Been Removed |
||||||||||||||
| symmetry | Space Group: P 21 21 21 | R_factor | |||||||||||||
| crystal
cell |
|
||||||||||||||
| method | X-Ray Diffraction | resolution | 1.9 Å | ||||||||||||
| ligand | CRO, MSE | ||||||||||||||
| number of helix | 4 | residue number | 236 | number of b-sheet | 11 | ||||||||||
| helix start and stop | |||||||||||||||
| Helix 1: From GLY 4 to PHE 8 Helix 2: From TRP 57 to LEU 60 Helix 3: From GIN 69 to PHE 71 Helix 4: From PHE 83 to SER 86 | |||||||||||||||
Show three different Rasmol pictures of GFP: whatever you like. highlight the chromophore by spacefill and green. a rotating molecule by GIF builder.
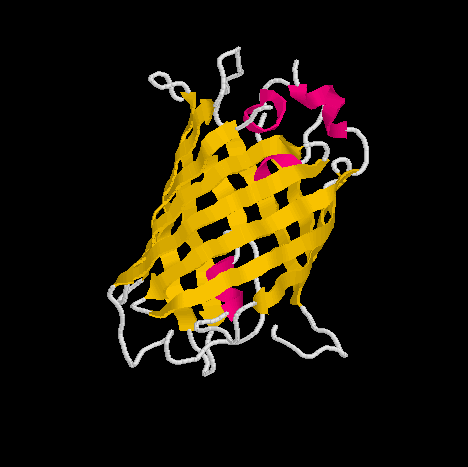

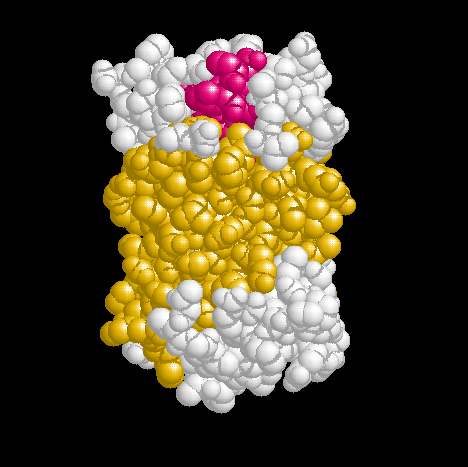
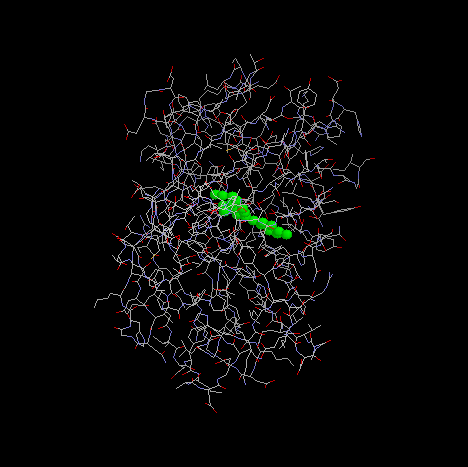
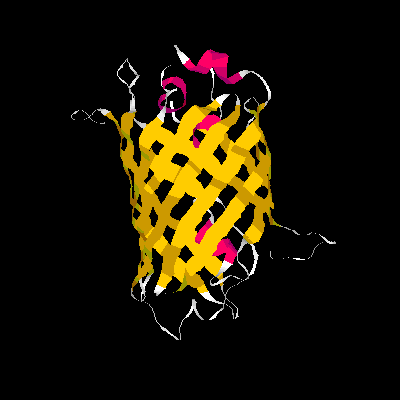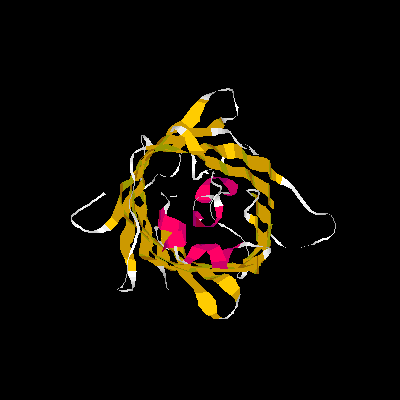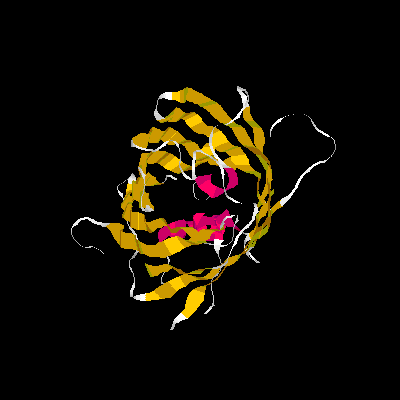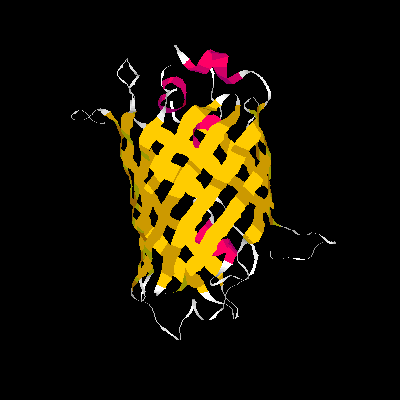
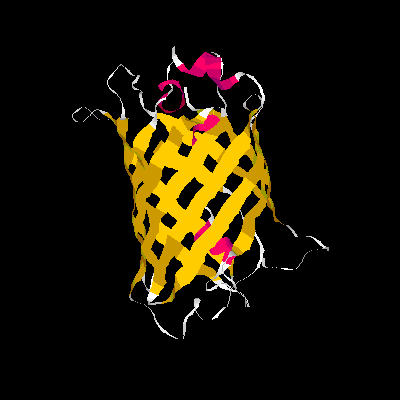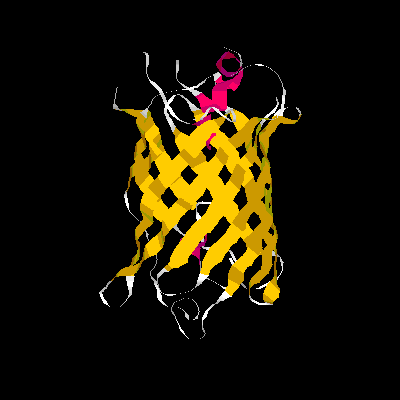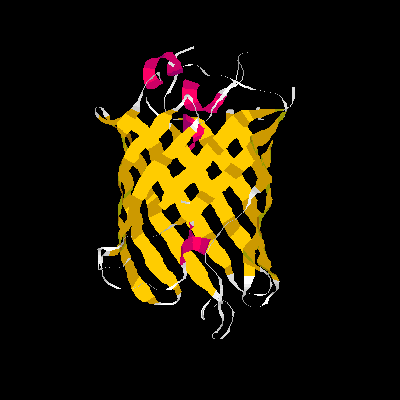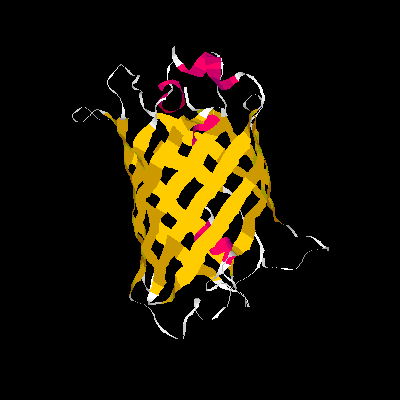
A blue emission variant of GFP has been made by mutation of two residues. Please find the structure of this blue emission variant. Show its structure and color the two mutated positions blue.
The mutation site : Y66H and Y145F
Back to Homework