David Liu, a student in the department of Lfe Sciences, got a clone from E coli.
Unfortunately, he doesn't know how to use the sequence analysis tools availabled in the
internet since he did not take Bioinformatics course before. Could you help him to do the
following analysis?
(1) Find its corresponding polypeptide sequence (DNA -> Protein translation).
(2) Identify this protein. Is it a new protein?
(3) Report the total number of negatively charged residues and positively charged residues.
(4) Color the protein by its charge.
(5) Draw the hydrophobicity map for this protein using Eisenberg hydrophobicity scale with window
size 7. The relative weight of the window edges compared to the window center should set to 40%
(6) Please help him to use Prosite scanning tool to find out possible functions or pattern of this
protein.
(7) Calculate its pI and molecular weight.
1.These sequence corresponding polypeptide sequence are shown as below from 5 to 3 frame
W M P C S G G N M K K S I L F I F L S V L S F S P F A Q D A K P V E S S K E K I T L E S K K C
N I A K K S N K S G P E S M N S S N Y C C E L C C N P A C T G C Y Stop Stop Y K G N Stop T V P
F I F V L I L M M S V T Y V L L L C Stop I N R
2.Blast the polypeptide sequence with NCBI Blast tool, it seems that it is
heat-stable enterotoxin STA4 precursor
3.The negatively charged residues are "DE" and positively charged residues are "k"
so there are 6 negatively charged residues and 10 positively charged residues in
this protein.
4.The negatively charged residuesare displaied in color red, positively charged
residues in purple, and the non-charged residues in black.
M P C S G G N M K K S I L F I F L S V L S F S P F A Q D A K P V E S S K E K I T L E S K K C
N I A K K S N K S G P E S M N S S N Y C C E L C C N P A C T G C Y
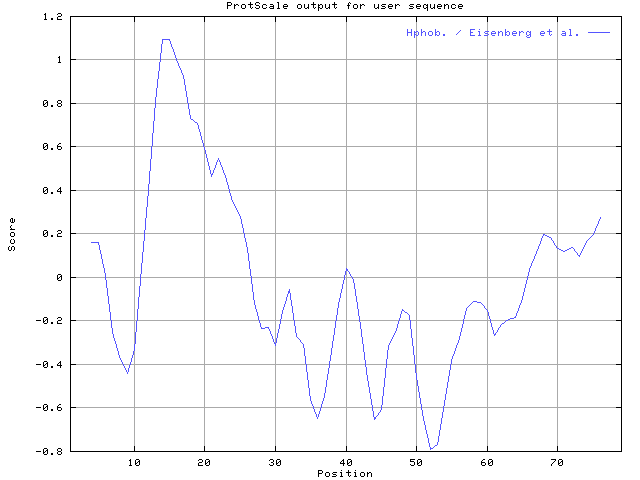
5.The hydrophobicity map displaied with window size 7, window center 40%
 6.One of the possible functions or pattern of this protein should be the
Heat-stable enterotoxins signature, and the ducumentation are shown below.
To see the other prosite results
Prosite:PDOC00246(ducumentation)
Prokaryotic heat-stable enterotoxins [1] are responsible for acute diarrhea.
The active toxin is a short peptide of around twenty residues which contains
six cysteines involved in three disulfide bonds, as shown in the following
schematic representation:
+-------+
+--|----+ |
| | | |
xxCCxxCCxxxCxxCxx
| |
+----+
'C': conserved cysteine involved in a disulfide bond.
We have taken the pattern of cysteines, along with three conserved residues,
as a signature pattern for this group of proteins
-Consensus pattern: C-C-x(2)-C-C-x-P-A-C-x-G-C
[The six C's are involved in disulfide bonds]
-Sequences known to belong to this class detected by the pattern: ALL.
-Other sequence(s) detected in SWISS-PROT: NONE.
-Last update: April 1990 / Pattern and text revised.
[ 1] Shimonishi Y., Hidaka Y., Koizumi M., Hane M., Aimoto S., Takeda T.,
Miwatani T., Takeda Y.
FEBS Lett. 215:165-170(1987).
7.The molecular weight and pI are shown as below:
Molecular weight: 8555.96
Theoretical pI: 8.66
6.One of the possible functions or pattern of this protein should be the
Heat-stable enterotoxins signature, and the ducumentation are shown below.
To see the other prosite results
Prosite:PDOC00246(ducumentation)
Prokaryotic heat-stable enterotoxins [1] are responsible for acute diarrhea.
The active toxin is a short peptide of around twenty residues which contains
six cysteines involved in three disulfide bonds, as shown in the following
schematic representation:
+-------+
+--|----+ |
| | | |
xxCCxxCCxxxCxxCxx
| |
+----+
'C': conserved cysteine involved in a disulfide bond.
We have taken the pattern of cysteines, along with three conserved residues,
as a signature pattern for this group of proteins
-Consensus pattern: C-C-x(2)-C-C-x-P-A-C-x-G-C
[The six C's are involved in disulfide bonds]
-Sequences known to belong to this class detected by the pattern: ALL.
-Other sequence(s) detected in SWISS-PROT: NONE.
-Last update: April 1990 / Pattern and text revised.
[ 1] Shimonishi Y., Hidaka Y., Koizumi M., Hane M., Aimoto S., Takeda T.,
Miwatani T., Takeda Y.
FEBS Lett. 215:165-170(1987).
7.The molecular weight and pI are shown as below:
Molecular weight: 8555.96
Theoretical pI: 8.66
Back to homework