- a)
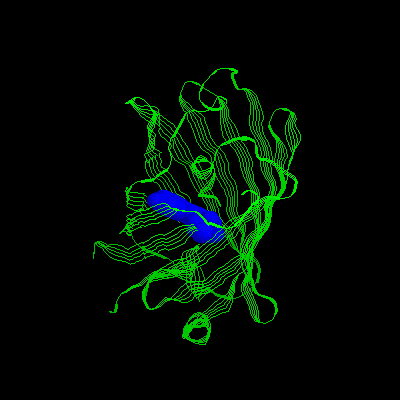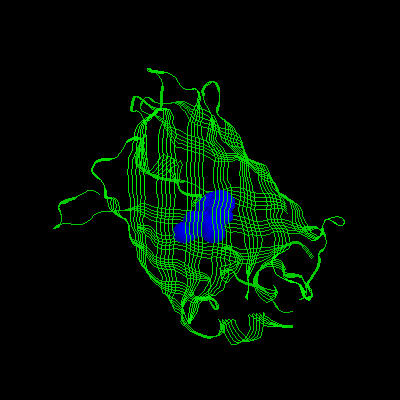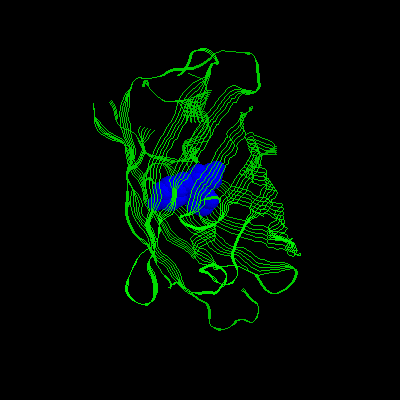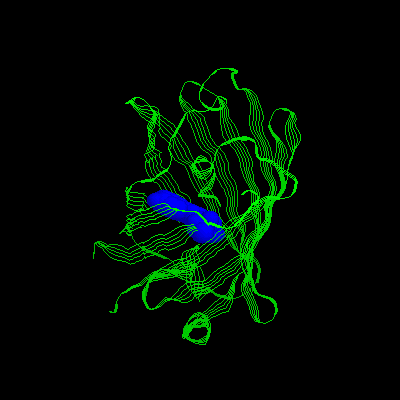
3.Mutation: S65, H66, And G67 Are Replaced
With Iic 66, Y145f
PC Lyu
HOMEWORK#5
Protein Data Bank
- due on 4/25
| PDB ID | 1EMA | 1GFL |
| Authers | M.Ormo, A.B.Cubitt, K.Kallio, L.A.Gross, R.Y.Tsien, S.J.Remington | F.Yang, L.G.Moss, G.N.Phillips Junior |
| Compound | Molecule: Green Fluorescent Protein
Chain: Null Engineered: Yes Mutation: 65 - 67 Replaced By Cro, Q80r Other_details: The Protein Contains Six Se-Methionines. Of These, The N-Terminal Met And Met 233 Are Not Present In The Entry |
Molecule: Green Fluorescent Protein
Chain: A, B Engineered: Yes Mutation: Q80r |
| Expression system | Organism_scientific: Aequorea Victoria
Organism_common: Jellyfish Plasmid: Prsetb (Invitrogen) Expression_system:Escherichia Coli Expression_system_strain: Jm109 (De3) Other_details: The N-Terminal His-Tag Has Been Removed |
Organism_scientific: Aequorea Victoria
Expression_system: Escherichia Coli Expression_system_plasmid: Ptu58 |
| Residue numbers | 708 | A chain:714 ;B chain:714 |
| NO. of heliex | 4 | A chain:6 ; B chain:6 |
| NO.of sheet | 11 | A chain:12 ; B chain:12 |
| The starting residue and ending residue of each helix. | HELIX 1: GLY 4- PHE 8
HELIX 2: TRP 57 - LEU 60 HELIX 3: GLN 69 - PHE 71 HELIX 4: PHE 83 - SER 86 |
A chain:
HELIX1:GLU 5-PHE 8 HELIX2:TRP 57 - PHE 64 HELIX3:GLN 69 - PHE71 HELIX 4:ASP 76 - HIS 81 HELIX 5:PHE 83-SER86 HELIX6:LYS156-LYS158 B chain: HELIX7:GLU5-PHE8 HELIX8:TRP57-PHE64 HELIX9:GLN69- PHE71 HELIX10:ASP76-HIS81 HELIX11: PHE83-SER86 HELIX12: LYS156-LYS158 |