HOMEWORK#5
Lymphoid enhancer-binding factor (LEF-1) and the
closely related T-cell factor 1 (TCF-1) are
sequence-specific and cell-type-specific DNA-binding
proteins that play important regulatory roles in
organogenesis and thymocyte differentiation. Please
find mouse LEF-1 and TCF-1 in SWISS-PROT and answer
the following questions:
1.Give their accession number.
LEF-1:P27782
TCF-1:Q00417
2.They share the same DNA-binding domain. What's
the name of this domain? What is the residue
number corresponding to the full-length protein?
LEF-1 HMG BOX:297-365
TCF-1 HMG BOX:188-256
3.Show the AA sequence of this DNA-binding
domain of each protein.
LEF-1
Lys Lys Pro Leu Asn Ala Phe Met Leu Tyr Met Lys Glu
Met Arg Ala Asn Val Val Ala Glu Cys Thr Leu Lys Glu
Ser Ala Ala Ile Asn Gln Ile Leu Gly Arg Arg Trp His
Ala Leu Ser Arg Glu Glu Gln Ala Lys Tyr Tyr Glu Leu
Ala Arg Lys Glu Arg Gln Leu His Met Gln Leu Tyr Pro
Gly Trp Ser
TCF-1
Ile Lys Lys Pro Leu Asn Ala Phe Met Leu Tyr Met Lys
Glu Met Arg Ala Lys Val Ile Ala Glu Cys Thr Leu Lys
Glu Ser Ala Ala Ile Asn Gln Ile Leu Gly Arg Arg Trp
His Ala Leu Ser Arg Glu Glu Gln Ala Lys Tyr Tyr Glu
Leu Ala Arg Lys Glu Arg Gln Leu His Met Gln Leu Tyr
Pro Gly Trp Ser
4.Calculate the pI/Mw and predict the secondary of
this DNA-binding domain of LEF-1.
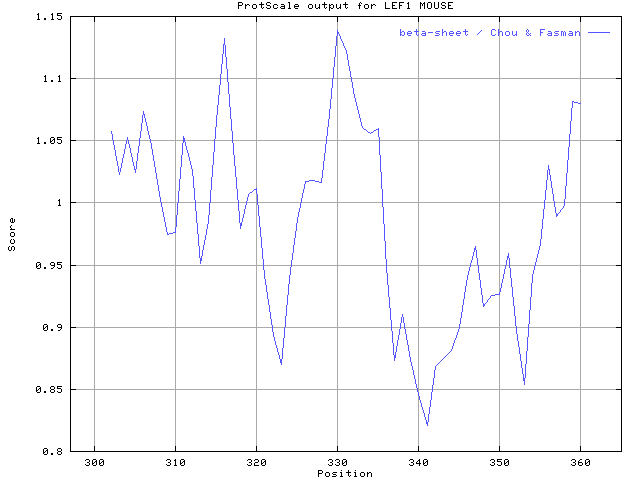
PI=9.66 MW=8239.65 dolton
Using the scale beta-sheet / Chou & Fasman,
the individual values for the 20 amino acids are:
1 2 3 4 5 6 7 8 9 10 11
0.50 0.51 0.54 0.59 0.72 1.00 0.72 0.59 0.54 0.51 0.50
edge center edge
 5.Calculate the hydropathicity ( Kyte & Doolittle) of
this DNA-binding domain of TCF-1.
1 2 3 4 5 6 7 8 9 10 11
0.50 0.60 0.70 0.80 0.90 1.00 0.90 0.80 0.70 0.60 0.50
edge center edge
5.Calculate the hydropathicity ( Kyte & Doolittle) of
this DNA-binding domain of TCF-1.
1 2 3 4 5 6 7 8 9 10 11
0.50 0.60 0.70 0.80 0.90 1.00 0.90 0.80 0.70 0.60 0.50
edge center edge
 6.Show the result of chymotrypsin cleavage of TCF-1.
( Displaying peptides with a mass bigger than 1000
Dalton with maximum number of missed cleavages= 1 )
mass position #MC peptide sequence
3987.117 262-297 1 GKKKRRSREKHQESTTGGKR NAFGTYPEKAAAPAPF
3456.738 14-46 1 MPYPPASGAGQHPQPQPPLH NKPGQPPHGVPQL
3392.775 256-284 1 SARDNYGKKKRRSREKHQES TTGGKRNAF
2874.486 54-80 1 SSPHPTPAPADISQKQGVHR PLQTPDL
2859.678 168-192 1 DRNLKTQAEPKAEKEAKKPV IKKPL
2791.453 140-167 1 GSGVPGHPAAIPHPAIVPSS GKQELQPY
2733.386 51-75 1 EHFSSPHPTPAPADISQKQG VHRPL
2693.571 172-195 1 KTQAEPKAEKEAKKPVIKKP LNAF
2686.472 262-284 0 GKKKRRSREKHQESTTGGKR NAF
2575.230 109-132 1 SPSCGYRQHFPAPTAAPGAP YPRF
2516.362 139-164 1 LGSGVPGHPAAIPHPAIVPS SGKQEL
2516.295 115-137 1 RQHFPAPTAAPGAPYPRFTH PSL
2403.278 140-164 0 GSGVPGHPAAIPHPAIVPSS GKQEL
2361.423 172-192 0 KTQAEPKAEKEAKKPVIKKP L
2320.216 54-75 0 SSPHPTPAPADISQKQGVHR PL
2171.222 203-222 1 RAKVIAECTLKESAAINQIL
2083.043 13-32 1 LMPYPPASGAGQHPQPQPPL
1981.019 115-132 0 RQHFPAPTAAPGAPYPRF
1978.969 87-105 1 TSGSMGQLPHTVSWPSPPL
1969.959 14-32 0 MPYPPASGAGQHPQPQPPL
1888.996 92-108 1 GQLPHTVSWPSPPLYPL
1802.966 33-49 1 HNKPGQPPHGVPQLSPL
1660.840 285-300 1 GTYPEKAAAPAPFLPM
1641.918 213-226 1 KESAAINQILGRRW
1515.795 92-105 0 GQLPHTVSWPSPPL
1505.797 33-46 0 HNKPGQPPHGVPQL
1491.802 200-212 1 KEMRAKVIAECTL
1331.670 227-237 1 HALSREEQAKY
1319.663 285-297 0 GTYPEKAAAPAPF
1228.538 252-261 1 YPGWSARDNY
1173.553 241-249 1 ARKERQLHM
1142.664 239-247 1 ELARKERQL
1103.624 203-212 0 RAKVIAECTL
1086.615 213-222 0 KESAAINQIL
1010.490 230-237 0 SREEQAKY
7.Show the result of Prosite scanning of LEF-1.
[1] PDOC00001 PS00001 ASN_GLYCOSYLATION
N-glycosylation site
Number of matches: 2
1 57-60 NESE
2 126-129 NGSL
[2] PDOC00005 PS00005 PKC_PHOSPHO_SITE
Protein kinase C phosphorylation site
Number of matches: 3
1 137-139 SNK
2 320-322 TLK
[3] PDOC00006 PS00006 CK2_PHOSPHO_SITE
Casein kinase II phosphorylation site
Number of matches: 7
1 40-43 SHPE
2 76-79 SSQE
3 157-160 TYSD
4 273-276 TDSD
5 320-323 TLKE
6 339-342 SREE
7 365-368 SARD
[4] PDOC00007 PS00007 TYR_PHOSPHO_SITE
Tyrosine kinase phosphorylation site
93-100 KHPDGGLY
[5] PDOC00008 PS00008 MYRISTYL
N-myristoylation site
Number of matches: 4
1 6-11 GGGGGG
2 7-12 GGGGGD
3 97-102 GGLYNK
4 166-171 GSHPSH
[6] PDOC00009 PS00009 AMIDATION
Amidation site
Number of matches: 2
1 331-334 LGRR
2 370-373 YGKK
6.Show the result of chymotrypsin cleavage of TCF-1.
( Displaying peptides with a mass bigger than 1000
Dalton with maximum number of missed cleavages= 1 )
mass position #MC peptide sequence
3987.117 262-297 1 GKKKRRSREKHQESTTGGKR NAFGTYPEKAAAPAPF
3456.738 14-46 1 MPYPPASGAGQHPQPQPPLH NKPGQPPHGVPQL
3392.775 256-284 1 SARDNYGKKKRRSREKHQES TTGGKRNAF
2874.486 54-80 1 SSPHPTPAPADISQKQGVHR PLQTPDL
2859.678 168-192 1 DRNLKTQAEPKAEKEAKKPV IKKPL
2791.453 140-167 1 GSGVPGHPAAIPHPAIVPSS GKQELQPY
2733.386 51-75 1 EHFSSPHPTPAPADISQKQG VHRPL
2693.571 172-195 1 KTQAEPKAEKEAKKPVIKKP LNAF
2686.472 262-284 0 GKKKRRSREKHQESTTGGKR NAF
2575.230 109-132 1 SPSCGYRQHFPAPTAAPGAP YPRF
2516.362 139-164 1 LGSGVPGHPAAIPHPAIVPS SGKQEL
2516.295 115-137 1 RQHFPAPTAAPGAPYPRFTH PSL
2403.278 140-164 0 GSGVPGHPAAIPHPAIVPSS GKQEL
2361.423 172-192 0 KTQAEPKAEKEAKKPVIKKP L
2320.216 54-75 0 SSPHPTPAPADISQKQGVHR PL
2171.222 203-222 1 RAKVIAECTLKESAAINQIL
2083.043 13-32 1 LMPYPPASGAGQHPQPQPPL
1981.019 115-132 0 RQHFPAPTAAPGAPYPRF
1978.969 87-105 1 TSGSMGQLPHTVSWPSPPL
1969.959 14-32 0 MPYPPASGAGQHPQPQPPL
1888.996 92-108 1 GQLPHTVSWPSPPLYPL
1802.966 33-49 1 HNKPGQPPHGVPQLSPL
1660.840 285-300 1 GTYPEKAAAPAPFLPM
1641.918 213-226 1 KESAAINQILGRRW
1515.795 92-105 0 GQLPHTVSWPSPPL
1505.797 33-46 0 HNKPGQPPHGVPQL
1491.802 200-212 1 KEMRAKVIAECTL
1331.670 227-237 1 HALSREEQAKY
1319.663 285-297 0 GTYPEKAAAPAPF
1228.538 252-261 1 YPGWSARDNY
1173.553 241-249 1 ARKERQLHM
1142.664 239-247 1 ELARKERQL
1103.624 203-212 0 RAKVIAECTL
1086.615 213-222 0 KESAAINQIL
1010.490 230-237 0 SREEQAKY
7.Show the result of Prosite scanning of LEF-1.
[1] PDOC00001 PS00001 ASN_GLYCOSYLATION
N-glycosylation site
Number of matches: 2
1 57-60 NESE
2 126-129 NGSL
[2] PDOC00005 PS00005 PKC_PHOSPHO_SITE
Protein kinase C phosphorylation site
Number of matches: 3
1 137-139 SNK
2 320-322 TLK
[3] PDOC00006 PS00006 CK2_PHOSPHO_SITE
Casein kinase II phosphorylation site
Number of matches: 7
1 40-43 SHPE
2 76-79 SSQE
3 157-160 TYSD
4 273-276 TDSD
5 320-323 TLKE
6 339-342 SREE
7 365-368 SARD
[4] PDOC00007 PS00007 TYR_PHOSPHO_SITE
Tyrosine kinase phosphorylation site
93-100 KHPDGGLY
[5] PDOC00008 PS00008 MYRISTYL
N-myristoylation site
Number of matches: 4
1 6-11 GGGGGG
2 7-12 GGGGGD
3 97-102 GGLYNK
4 166-171 GSHPSH
[6] PDOC00009 PS00009 AMIDATION
Amidation site
Number of matches: 2
1 331-334 LGRR
2 370-373 YGKK
¦^¤W¤@¶



