![]() Homework
5
Homework
5
Q: Lymphoid enhancer-binding factor (LEF-1) and the closely related T-cell factor 1 (TCF-1) are sequence-specific and cell-type-specific DNA-binding proteins that play important regulatory roles in organogenesis and thymocyte differentiation. Please find mouse LEF-1 and TCF-1 in SWISS-PROT and answer the following questions:
Q1: Give their accession number.
Ans:
LEF-1 accession number : p27782
TCF-1 accession number : Q00417
Q2: They share the same DNA-binding domain. What's the name of this domain? What is the residue number corresponding to the full-length protein?
Ans: (1) HMG BOX
(2) LEF-1 DNA binding domain residue number 297-365
TCF-1 DNA binding domain residue number 188-256
Q3: Show the AA sequence of this DNA-binding domain of each protein.
Ans:
LEF-1 : IKKPLNAFML YMKEMRANVV AECTLKESAA INQILGRRWH ALSREEQAKY YELARKERQL HMQLYPGWS
TCF-1 : IKKPLNAFML YMKEMRAKVI AECTLKESAA INQILGRRWH ALSREEQAKY YELARKERQL HMQLYPGWS
Q4: Calculate the pI/Mw and predict the secondary of this DNA-binding domain of LEF-1.
Ans: Theoretical pI: 9.66
Molecular weight: 8239.6
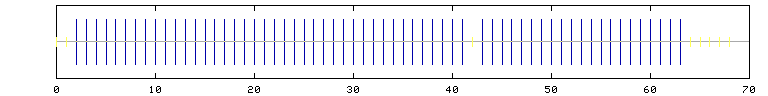
Hierarchical Neural Network-result for DNA-Binding Domain
10 20 30 40 50 60 70 80
| | | | | | | |
IKKPLNAFMLYMKEMRAKVIAECTLKESAAINQILGRRWHALSREEQAKYYELARKERQLHMQLYPGWS
cchhhhhhhhhhhhhhhhhhhhhhhhhhhhhhhhhhhhhhhhchhhhhhhhhhhhhhhhhhhhhccccc
Q5: Calculate the hydropathicity ( Kyte & Doolittle) of this DNA-binding domain of TCF-1.
Ans:
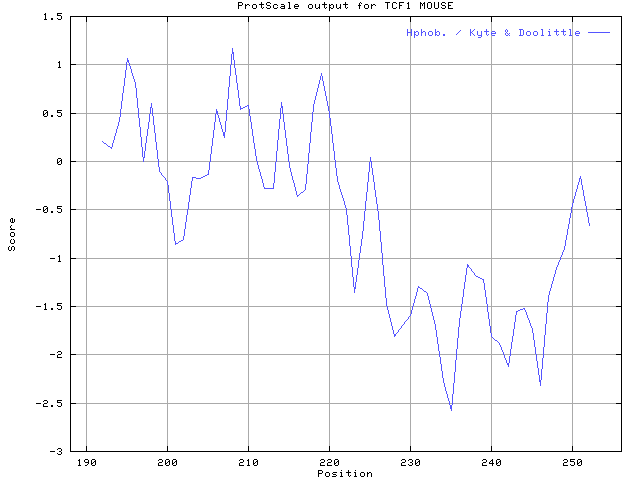
¡@
Q6: Show the result of chymotrypsin cleavage of TCF-1. ( Displaying peptides with a mass bigger than 1000 Dalton with maximum number of missed cleavages = 1 )
Ans:
[Theoretical pI: 9.79 / Mw (average mass): 33608.54 / Mw (monoisotopic mass): 33587.19]
| mass | position | #MC | peptide sequence |
| 3987.117 | 262-297 |
1 |
GKKKRRSREKHQESTTGGKR NAFGTYPEKAAAPAPF |
| 3456.738 | 14-46 |
1 |
MPYPPASGAGQHPQPQPPLH NKPGQPPHGVPQL |
| 3392.775 | 256-284 |
1 |
SARDNYGKKKRRSREKHQES TTGGKRNAF |
| 2874.486 | 54-80 |
1 |
SSPHPTPAPADISQKQGVHR PLQTPDL |
| 2859.678 | 168-192 |
1 |
DRNLKTQAEPKAEKEAKKPV IKKPL |
| 2791.453 | 140-167 |
1 |
GSGVPGHPAAIPHPAIVPSS GKQELQPY |
| 2733.386 | 51-75 |
1 |
EHFSSPHPTPAPADISQKQG VHRPL |
| 2693.571 | 172-195 |
1 |
KTQAEPKAEKEAKKPVIKKP LNAF |
| 2686.472 | 262-284 |
0 |
GKKKRRSREKHQESTTGGKR NAF |
| 2575.230 | 109-132 |
1 |
SPSCGYRQHFPAPTAAPGAP YPRF |
| 2516.362 | 139-164 |
1 |
LGSGVPGHPAAIPHPAIVPS SGKQEL |
| 2516.295 | 115-137 |
1 |
RQHFPAPTAAPGAPYPRFTH PSL |
| 2403.278 | 140-164 |
0 |
GSGVPGHPAAIPHPAIVPSS GKQEL |
| 2361.423 | 172-192 |
0 |
KTQAEPKAEKEAKKPVIKKP L |
| 2320.216 | 54-75 |
0 |
SSPHPTPAPADISQKQGVHR PL |
| 2171.222 | 203-222 |
1 |
RAKVIAECTLKESAAINQIL |
| 2083.043 | 13-32 |
1 |
LMPYPPASGAGQHPQPQPPL |
| 1981.019 | 115-132 |
0 |
RQHFPAPTAAPGAPYPRF |
| 1978.969 | 87-105 |
1 |
TSGSMGQLPHTVSWPSPPL |
| 1969.959 | 14-32 |
0 |
MPYPPASGAGQHPQPQPPL |
| 1888.996 | 92-108 |
1 |
GQLPHTVSWPSPPLYPL |
| 1802.966 | 33-49 |
1 |
HNKPGQPPHGVPQLSPL |
| 1660.840 | 285-300 |
1 |
GTYPEKAAAPAPFLPM |
| 1641.918 | 213-226 |
1 |
KESAAINQILGRRW |
| 1515.795 | 92-105 |
0 |
GQLPHTVSWPSPPL |
| 1505.797 | 33-46 |
0 |
HNKPGQPPHGVPQL |
| 1491.802 | 200-212 |
1 |
KEMRAKVIAECTL |
| 1331.670 | 227-237 |
1 |
HALSREEQAKY |
| 1319.663 | 285-297 |
0 |
GTYPEKAAAPAPF |
| 1228.538 | 252-261 |
1 |
YPGWSARDNY |
| 1173.553 | 230-238 |
1 |
SREEQAKYY |
| 1168.637 | 241-249 |
1 |
ARKERQLHM |
| 1142.664 | 239-247 |
1 |
ELARKERQL |
| 1103.624 | 203-212 |
0 |
RAKVIAECTL |
| 1086.615 | 213-222 |
0 |
KESAAINQIL |
| 1010.490 | 230-237 |
0 |
SREEQAKY |
¡@
Q7: Show the result of Prosite scanning of LEF-1.
Ans: Scan of LEF1_MOUSE (P27782)
LYMPHOID ENHANCER BINDING FACTOR 1.
MUS MUSCULUS (MOUSE).
¡@
¡@
[1] PDOC00001 PS00001 ASN_GLYCOSYLATION
N-glycosylation site
Number of matches: 2
1 57-60 NESE
2 126-129 NGSL
¡@
[2] PDOC00005 PS00005 PKC_PHOSPHO_SITE
Protein kinase C phosphorylation site
Number of matches: 3
1 137-139 SNK
2 320-322 TLK
3 365-367 SAR
¡@
[3] PDOC00006 PS00006 CK2_PHOSPHO_SITE
Casein kinase II phosphorylation site
Number of matches: 7
1 40-43 SHPE
2 76-79 SSQE
3 157-160 TYSD
4 273-276 TDSD
5 320-323 TLKE
6 339-342 SREE
7 365-368 SARD
¡@
[4] PDOC00007 PS00007 TYR_PHOSPHO_SITE
Tyrosine kinase phosphorylation site
93-100 KHPDGGLY
¡@
[5] PDOC00008 PS00008 MYRISTYL
N-myristoylation site
Number of matches: 4
1 6-11 GGGGGG
2 7-12 GGGGGD
3 97-102 GGLYNK
4 166-171 GSHPSH
¡@
[6] PDOC00009 PS00009 AMIDATION
Amidation site
Number of matches: 2
1 331-334 LGRR
2 370-373 YGKK