Overview
The interactive program chemprop is designed to aid the user in examining the properties of a receptor in the vicinity of a small molecule that has been oriented using dock. The program takes as input a receptor coordinate file, a file containing the coordinates of one or more ligands, and several parameters, described below, depending on the option chosen. There are two options: electrostatics and hydrogen bonding.
In the electrostatic option, the electrostatic potential from the receptor is calculated at each ligand molecule atom center. The electrostatic potential at the position of ligand atom j, ej, is calculated using equation 1, where qi is the partial charge on the receptor atom i, D is the dielectric constant, and rij is the distance between atoms i and j:
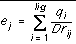 Equation 11
Equation 11
Only the receptor atoms contribute to the value of the electrostatic potential. The partial atomic charges used are those from the amber united-atom force field (Weiner, et al., 1984). Only standard amino acid residues can be accommodated by this program. The receptor file must include the hydrogen atoms attached to nitrogens, hydroxyl oxygens, and sulfurs, and the lone pairs on the sulfurs. A new pdb-format coordinate file is written out for the ligand molecules, where the electrostatic potential is printed in the temperature factor column. The molecules can then be displayed using a molecular graphics package and colored according to electrostatic potential.
The hydrogen bond option helps the user identify places on the ligands where it might be appropriate to design in a group capable of hydrogen bonding to the receptor. Potential hydrogen bond positions are identified as any ligand atom within a user-specified distance of a receptor nitrogen or oxygen atom. This option was intended mainly for use with ligand heavy atom (nonhydrogen) coordinates only. Two files are output: a coordinate file, and a file listing the potential hydrogen bonds for each ligand in the input file. In the coordinate file, each potential hydrogen bond is written in pdb format as a residue with two atoms. One atom is located at the receptor nitrogen or oxygen and the other is located at the ligand atom. The residues are separated by ter cards. The residue and atom names depend on the protein atom. The residue is named acc if the protein atom is a carbonyl oxygen, dnr if the protein atom is an amide or amine nitrogen, and doa if the protein atom is a hydroxyl oxygen or a histidine side chain nitrogen. The receptor and ligand atoms in an acc residue are named a and d, respectively; in a dnr residue they are named d and a, respectively; and in a doa residue they are named e1 and e2, respectively. Viewing these possible hydrogen bonds with the receptor and ligand is useful for design purposes and assessing whether the angles are consistent with strong hydrogen bonding.