4.4.1 CoMSIA Field Descriptions
In both CoMFA and CoMSIA, a group of structurally aligned molecules are represented in terms of fields around the molecule. These molecular property fields are evaluated between a probe atom and each molecule, at regularly spaced intervals on a grid. The value displayed in the MSS for each CoMSIA column is the RMS deviation of the points that constitute the field.
CoMFA calculates steric fields using a Lennard-Jones potential, and electrostatic fields using a Coulombic potential [Ref. 28]. While this approach has been widely accepted and exceptionally valuable, it is not without problems. In particular, both potential functions are very steep near the van der Waals surface of the molecule, causing rapid changes in surface descriptions, and requiring the use of cut-off values so calculations are not done inside the molecular surface. In addition, a scaling factor is applied to the steric field, so both fields can be used in the same PLS analysis. Finally, changes in orientation of the superimposed molecule set, relative to the calculation grid, can cause significant changes in CoMFA results, again probably due to strict cut-off values.
In CoMSIA, five different similarity fields are calculated: steric, electrostatic, hydrophobic, hydrogen bond donor and hydrogen bond acceptor. These fields were selected to cover the major contributions to ligand binding [Ref. 29]. Similarity indices are calculated at regularly spaced grid points for the pre-aligned molecules. A comparison of the relative shapes of CoMSIA and CoMFA fields is shown below.
Figure 20 Shapes of various functions.
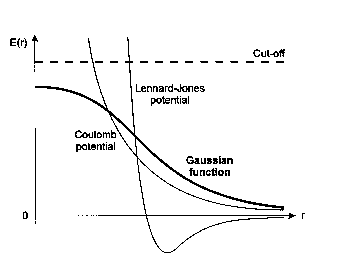
For the distance dependence between the probe atom and the molecule atoms a Gaussian function is used. Because of the different shape of the Gaussian function, the similarity indices can be calculated at all grid points, both inside and outside the molecular surface.
The equation used to calculate the similarity indices is as follows:
[EQ 4] 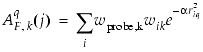
- A is the similarity index at grid point q, summed over all atoms i of the molecule j under investigation.
- wprobe, k is the probe atom with radius 1 Å, charge +1, hydrophobicity +1, hydrogen bond donating +1, hydrogen bond accepting +1.
- wik is the actual value of the physicochemical property k of atom i.
- riq is the mutual distance between the probe atom at grid point q and atom i of the test molecule.
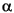 is the attenuation factor, with a default value of 0.3, and an optimal value normally between 0.2 and 0.4 [Ref. 31]. Larger vales result in a steeper Gaussian function, and a strong attenuation of the distance-dependent effects of molecular similarity. Global molecular features become less important, and there is little averaging of local features. With an
is the attenuation factor, with a default value of 0.3, and an optimal value normally between 0.2 and 0.4 [Ref. 31]. Larger vales result in a steeper Gaussian function, and a strong attenuation of the distance-dependent effects of molecular similarity. Global molecular features become less important, and there is little averaging of local features. With an 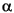 of 0.3, each property value of a given atom is felt by 74.1% at 1 Å from the atom, by 30.1% at 2 Å, and by 6.7% at 3 Å.
of 0.3, each property value of a given atom is felt by 74.1% at 1 Å from the atom, by 30.1% at 2 Å, and by 6.7% at 3 Å.
The actual values of the physicochemical properties for each atom are based on the following sources:
- Steric CoMSIA field: directly from the VDW table in $TA_ASCTABLES/ATOM_DEF. However the values are internally coded and cannot be modified.
- Electrostatic CoMSIA fields: uses the current charges calculated for the molecule. These values are accessed at the time the field is computed and thus can be modified by recalculating or directly editing the charge.
- Hydrogen bond donor and acceptor fields: To create the donor and acceptor fields, CoMSIA first creates dummy atoms at donor and acceptor sites much like DISCO's extension points [Ref. 29]. Each of these points is used for either the donor and acceptor field. Any donor or acceptor site points within 1.8 Å of a real atom in the molecule is ignored in the field calculation. The location of these donor and acceptor points may be seen using the expression generator %comsia_info().
Note: The Tripos implementation of CoMSIA uses a nomenclature opposite to that used in Ref. 29 and Ref. 32, in accordance with modifications made by the original authors.
- The acceptor field contains information about where hydrogen bond donating groups should be on the receptor.
- The donor field describes where hydrogen bond acceptor groups should be located on the receptor.
- Hydrophobic CoMSIA fields: the atomic values are directly based on the research of Viswanadhan et.al [Ref. 33]. The values are stored in the table $TA_MOLTABLES/comsia/data.lip.
Note: The SYBYL implementation of CoMSIA has changed the sign of the fields from negative to positive values in order to conform to the conventions of standard CoMFA graphs. This sign change does not alter the results. Knowledge of this sign change is useful when evaluating published results with inverted sign.
The end result of a QSAR analysis is often a set of contour maps, showing which areas around the molecule interact favorably (or unfavorably) with the surrounding environment. While using more fields does not necessarily increase the accuracy or predictive ability of the model (because of the correlations between fields), it can make the contour maps easier to interpret by enabling partitioning of variance into the different field types. The use of Gaussian functions also generally results in smoother, less fragmented surfaces.


Copyright © 1999, Tripos Inc. All rights
reserved.







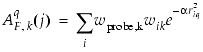
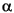 is the attenuation factor, with a default value of 0.3, and an optimal value normally between 0.2 and 0.4 [Ref. 31]. Larger vales result in a steeper Gaussian function, and a strong attenuation of the distance-dependent effects of molecular similarity. Global molecular features become less important, and there is little averaging of local features. With an
is the attenuation factor, with a default value of 0.3, and an optimal value normally between 0.2 and 0.4 [Ref. 31]. Larger vales result in a steeper Gaussian function, and a strong attenuation of the distance-dependent effects of molecular similarity. Global molecular features become less important, and there is little averaging of local features. With an 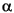 of 0.3, each property value of a given atom is felt by 74.1% at 1 Å from the atom, by 30.1% at 2 Å, and by 6.7% at 3 Å.
of 0.3, each property value of a given atom is felt by 74.1% at 1 Å from the atom, by 30.1% at 2 Å, and by 6.7% at 3 Å. 
