Key for assignment 7
1. Search human Mlh1 or its homologous proteins in Protein Data Bank (PDB) database. You just need to search
for protein structure, do not find complex structure. Give its (a) ID number,
(b) Name of the molecule, (c) experimental method for structure determination
and (d) Resolution of the structure.
(b) MutL
(c) X-RAY DIFFRACTION
(d) 2.9 ANGSTROMS
PDB
-> click on SearchLite -> type
MutL -> hit Search button -> select 1BKN -> click EXPLORE
OR
Go to Protein Structure Resource (PSR) -> hit BLAST PDB button -> paste Mlh1
sequence into the box
-> hit Search button -> You will find
1BKN on the top of the search results -> search for 1BKN in
PDB
2. Show its structure in ribbons form (400x400).
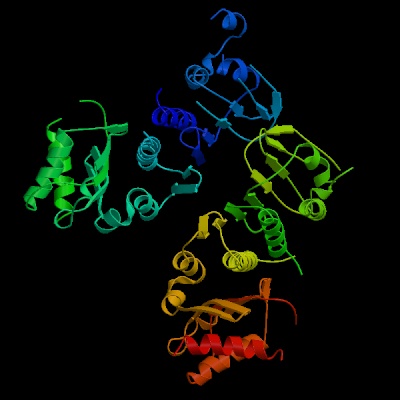
Click on View
Structure -> Go to "Custom Size Images:" -> type 400 in the box
of size ->choose JPEG format -> hit Create
image button
3. Give the structural informations of this PDB file (a) number of protein
chains, (b) number of helix in one protein chain, (c) the first and last
residues in the helix 6 .
(a) 2
(b) 11
(c) Thr 166 - Ala
181
4. Give the Class, Fold, Superfamily and Family of this protein in
Structural Classification of Proteins (SCOP).
1. Class: Alpha and beta proteins
(a+b)
Mainly antiparallel beta
sheets (segregated alpha and beta regions)
2. Fold: ATPase domain of HSP90 chaperone/DNA
topoisomerase II/histidine kinase
8-stranded mixed
beta-sheet; 2 layers: alpha/beta
3. Superfamily: ATPase domain of HSP90 chaperone/DNA
topoisomerase II/histidine kinase
4. Family: DNA gyrase/MutL, N-terminal
domain
5. (a) Show Ramachandran plot for this PDB file.
(b) How many residues are in the most favoured regions of Ramachandran
plot?
(a)
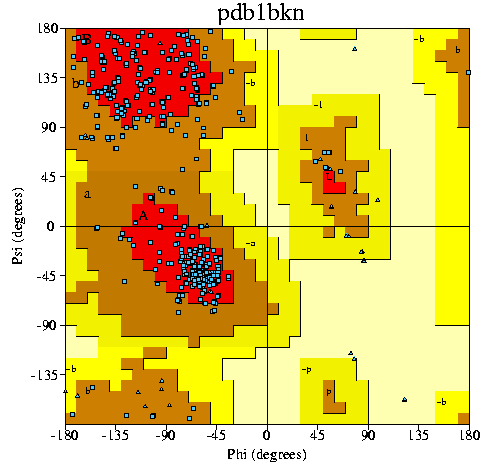
(b) 379 residues (80.5%) are in the most favoured
regions.
PCL 12/02/2001