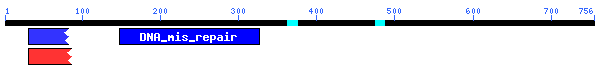
Compare human colon cancer gene MLH1 with other genes.
- To use ORF finder to translate DNA sequence to protein sequence in all reading
frames. -
- To use blastn, blastp, CD search and blast 2 sequence programs for searching
and comparison. -
-Deadline- 11/06/2001
1. Compare MLH1 (answer of assignment 2.6) and mutS (answer of 2.7) sequence.
From the homepage of the NCBI, we can find "Tools" at the left. We keep going into the BLAST system and choose the "Standard nucleatide-nucleartide BLAST". Get in to the "Blast 2 sequences" and pasted the two sequences on.
But finally we can only get no answer...
Sequence 1 lcl|seq_1 Length 2484
Sequence 2 lcl|seq_2 Length 2947
No significant similarity was found
2. Translate the above two gene sequences to protein sequences.
We copy the sequence, and past it on the Nucleotide query of the Translated BLAST Research. After the BLAST action and Format action after that:
MHL1
BLASTX 2.2.1 [Apr-13-2001]
Reference:
Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
RID: 1004725957-17574-30390
Query=
(2484 letters)
Database: nr
791,492 sequences; 251,575,206 total letters
Sequences producing significant alignments: (bits) Value
gi|4557757|ref|NP_000240.1| mutL homolog 1; mutL
(E. coli) ... 1443 0.0
gi|466462|gb|AAA17374.1| (U07418) human homolog of E. coli ... 1442 0.0
gi|604369|gb|AAA85687.1| (U17857) hMLH1 gene product [Homo ... 1429 0.0
gi|13878583|sp|Q9JK91|MLH1_MOUSE DNA MISMATCH REPAIR PROTEI... 1290 0.0
gi|13591989|ref|NP_112315.1| mismatch repair protein [Rattu... 1263 0.0
>gi|4557757|ref|NP_000240.1| mutL homolog 1; mutL (E. coli) homolog 1 [Homo
sapiens]
MSFVAGVIRRLDETVVNRIAAGEVIQRPANAIKEMIENCLDAKSTSIQVIVKEGGLKLIQIQDNGTGIRKEDLDIVCERF
TTSKLQSFEDLASISTYGFRGEALASISHVAHVTITTKTADGKCAYRASYSDGKLKAPPKPCAGNQGTQITVEDLFYNIA
TRRKALKNPSEEYGKILEVVGRYSVHNAGISFSVKKQGETVADVRTLPNASTVDNIRSIFGNAVSRELIEIGCEDKTLAF
KMNGYISNANYSVKKCIFLLFINHRLVESTSLRKAIETVYAAYLPKNTHPFLYLSLEISPQNVDVNVHPTKHEVHFLHEE
SILERVQQHIESKLLGSNSSRMYFTQTLLPGLAGPSGEMVKSTTSLTSSSTSGSSDKVYAHQMVRTDSREQKLDAFLQPL
SKPLSSQPQAIVTEDKTDISSGRARQQDEEMLELPAPAEVAAKNQSLEGDTTKGTSEMSEKRGPTSSNPRKRHREDSDVE
MVEDDSRKEMTAACTPRRRIINLTSVLSLQEEINEQGHEVLREMLHNHSFVGCVNPQWALAQHQTKLYLLNTTKLSEELF
YQILIYDFANFGVLRLSEPAPLFDLAMLALDSPESGWTEEDGPKEGLAEYIVEFLKKKAEMLADYFSLEIDEEGNLIGLP
LLIDNYVPPLEGLPIFILRLATEVNWDEEKECFESLSKECAMFYSIRKQYISEESTLSGQQSEVPGSIPNSWKWTVEHIV
YKALRSHILPPKHFTEDGNILQLANLPDLYKVFERC
E.coli mismatch repair gene mitS:
BLASTX 2.2.1 [Apr-13-2001]
Reference:
Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
RID: 1004725247-10838-22729
Query=
(2947 letters)
Database: nr
791,492 sequences; 251,575,206 total letters
Sequences producing significant alignments: (bits) Value
gi|4557761|ref|NP_000242.1| mutS homolog 2; mutS
(E. coli) ... 1778 0.0
gi|2135744|pir||I37550 mismatch repair protein MSH2 - human... 1675 0.0
gi|6678938|ref|NP_032654.1| mutS homolog 2 (E. coli) [Mus m... 1658 0.0
gi|726086|gb|AAA75027.1| (U21011) MutS homolog 2 [Mus muscu... 1657 0.0
gi|13591999|ref|NP_112320.1| mismatch repair protein [Rattu... 1646 0.0
gi|7448104|pir||I64827 gene MSH2 protein - human >gi|100088... 1503 0.0
gi|1079288|pir||S53609 DNA mismatch repair protein MSH2 - A... 1451 0.0
gi|14736868|ref|XP_034901.1| mutS homolog 2 [Homo sapiens] ... 1334 0.0
gi|15625578|gb|AAL04169.1|AF412833_1 (AF412833) mismatch re... 1321 0.0
gi|1000883|gb|AAB59571.1| (L47579) Insertion mutation resul... 1042 0.0
>gi|4557761|ref|NP_000242.1| mutS homolog 2; mutS (E. coli) homolog 2 [Homo
sapiens]
MAVQPKETLQLESAAEVGFVRFFQGMPEKPTTTVRLFDRGDFYTAHGEDALLAAREVFKTQGVIKYMGPAGAKNLQSVVL
SKMNFESFVKDLLLVRQYRVEVYKNRAGNKASKENDWYLAYKASPGNLSQFEDILFGNNDMSASIGVVGVKMSAVDGQRQ
VGVGYVDSIQRKLGLCEFPDNDQFSNLEALLIQIGPKECVLPGGETAGDMGKLRQIIQRGGILITERKKADFSTKDIYQD
LNRLLKGKKGEQMNSAVLPEMENQVAVSSLSAVIKFLELLSDDSNFGQFELTTFDFSQYMKLDIAAVRALNLFQGSVEDT
TGSQSLAALLNKCKTPQGQRLVNQWIKQPLMDKNRIEERLNLVEAFVEDAELRQTLQEDLLRRFPDLNRLAKKFQRQAAN
LQDCYRLYQGINQLPNVIQALEKHEGKHQKLLLAVFVTPLTDLRSDFSKFQEMIETTLDMDQVENHEFLVKPSFDPNLSE
LREIMNDLEKKMQSTLISAARDLGLDPGKQIKLDSSAQFGYYFRVTCKEEKVLRNNKNFSTVDIQKNGVKFTNSKLTSLN
EEYTKNKTEYEEAQDAIVKEIVNISSGYVEPMQTLNDVLAQLDAVVSFAHVSNGAPVPYVRPAILEKGQGRIILKASRHA
CVEVQDEIAFIPNDVYFEKDKQMFHIITGPNMGGKSTYIRQTGVIVLMAQIGCFVPCESAEVSIVDCILARVGAGDSQLK
GVSTFMAEMLETASILRSATKDSLIIIDELGRGTSTYDGFGLAWAISEYIATKIGAFCMFATHFHELTALANQIPTVNNL
HVTALTTEETLTMLYQVKKGVCDQSFGIHVAELANFPKHVIECAKQKALELEEFQYIGESQGYDIMEPAAKKCYLEREQG
EKIIQEFLSKVKQMPFTEMSEENITIKLKQLKAEVIAKNNSFVNEIISRIKVTT
3.Perform protein sequence homology searching for MLH1 in GenBank. Give the
10 highest hits.
Click the Protein query - Translated db [tblastn] of Translated BLAST Research and past the protein sequence. After the BLAST action and Format action, the reply will be like this:
Reference:
Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
RID: 1004726621-24036-23745
Query=
(756 letters)
Database: nt
1,000,461 sequences; 70,461,447 total letters
Sequences producing significant alignments: (bits) Value
gi|14725770|ref|XM_044891.1| Homo sapiens mutL (E. coli) ho... 1443 0.0
gi|13905125|gb|BC006850.1|BC006850 Homo sapiens, mutL (E. c... 1443 0.0
gi|4557756|ref|NM_000249.1| Homo sapiens mutL (E. coli) hom... 1443 0.0
gi|463988|gb|U07343.1|HSU07343 Human DNA mismatch repair pr... 1443 0.0
gi|466461|gb|U07418.1|HSHMLHI Human DNA mismatch repair (hm... 1442 0.0
gi|7595953|gb|AF250844.1|AF250844 Mus musculus MutL homolog... 1290 0.0
gi|13591988|ref|NM_031053.1| Rattus norvegicus mismatch rep... 1263 0.0
gi|1724117|gb|U80054.1|RNU80054 Rattus norvegicus mismatch ... 1263 0.0
gi|12835157|dbj|AK004105.1|AK004105 Mus musculus 18 days em... 771 0.0
gi|13543416|gb|BC005866.1|BC005866 Homo sapiens, Similar to... 738 0.0
4. Compare human MLH1 protein with MLH1 in M. musculus, R. norvegicus and D.
melanogaster. Give the pairwise alignment and % of sequence smility.
We click the Standard Protein-protein BLAST of the Protein BLAST. We paste the amino acid sequence, and then format, we can then find information below:
gi|7595954|gb|AAF64514.1|AF250844_1 (AF250844) MutL homolog 1 protein [Mus
musculus]
Length = 760
Score = 1292 bits (3344), Expect = 0.0
Identities = 651/760 (85%), Positives = 693/760 (90%),
Gaps = 4/760 (0%)
>gi|13591989|ref|NP_112315.1| mismatch repair protein [Rattus
norvegicus]
gi|13878571|sp|P97679|MLH1_RAT DNA MISMATCH REPAIR PROTEIN MLH1 (MUTL PROTEIN
HOMOLOG 1)
gi|1724118|gb|AAB38506.1| (U80054) mismatch repair protein [Rattus
norvegicus]
Length = 757
Score = 1289 bits (3336), Expect = 0.0
Identities = 639/758 (84%), Positives = 684/758 (89%),
Gaps = 3/758 (0%)
>gi|7304079|gb|AAF59117.1| (AE003838) Mlh1 gene product [Drosophila
melanogaster]
Length = 664
Score = 615 bits (1586), Expect = e-175
Identities = 335/751 (44%), Positives = 453/751 (59%),
Gaps = 94/751 (12%)
5. Search the conserve domain (CD) for MLH1. Give the position of the CD, name
of CD and Pfam ID number.
Click the Search the Conserved Domain Database using RPS-BLAST, then paste the protein sequence and search:
RPS-BLAST 2.2.1 [Aug-1-2001]
Query= local sequence:
(756 letters)
Database: oasis_sap.v1.54
3693 PSSMs; 718,011 total columns
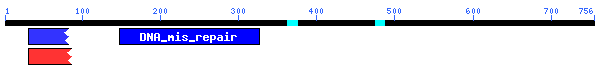
PSSMs producing significant alignments: Score
gnl|Pfam|pfam01119 DNA_mis_repair, DNA mismatch repair protein. Also known as
the... 202 7e-53
gnl|Pfam|pfam02518 HATPase_c, Histidine kinase-, DNA gyrase B-, phytochrome-like
... 40.8 3e-04
gnl|Smart|smart00387 HATPase_c, Histidine kinase-like ATPases; Histidine kinase-,
D... 40.0 5e-04
6. Show multiple alignment of MLH1 conserve domain with 5 sequences from the
top of the CD alignment.
We choose the one:
gnl|Pfam|pfam01119, DNA_mis_repair, DNA mismatch repair protein. Also known
as the mutL/hexB/PMS1 family.
We change the query to multiple alignment to displaying up to 5 sequence from the top of the CD alignment. The result is like this:
10 20 30 40 50 60
....*....|....*....|....*....|....*....|....*....|....*....|
consensus 1 GTTVEVRDLFYNLPVRRKFLKSPKKEFRKILDLLQRYALIHPNVSFSLTKEG--KALLQL 58
query 147 GTQITVEDLFYNIATRRKALKNPSEEYGKILEVVGRYSVHNAGISFSVKKQG--ETVADV 204
1B63_A 144 GTTLEVLDLFYNTPARRKFLRTEKTEFNHIDEIIRRIALARFDVTINLSHNG--KIVRQY 201
gi 8039787 159 GTVVRVEQLFENFPARKRFLGRQSAETTLCRSALIDVSLAHHPVEFRFTVDGthKLTLLS
218
gi 8928214 141 GTIVDVTKIFHNFPARKRFLKQEPIETKMCLKVLEEKIITHPEINFEIN-LN--QKLRKI
197
70 80 90 100 110 120
....*....|....*....|....*....|....*....|....*....|....*....|
consensus 59 KTSP--S-SLKERIRSVFGTAVLKNLIPF--EEKDGDFRIEG-FISSPNVSR-SSRDRQF 111
query 205 RTLP--NaSTVDNIRSIFGNAVSRELIEIgcEDKTLAFKMNG-YISNANYSV--KKCIFL 259
1B63_A 202 RAVPegG-QKERRLGAICGTAFLEQALAI--EWQHGDLTLRG-WVADPNHTTpALAEIQY 257
gi 8039787 219 QQTR--K-DRCLETQMLKGDPALFHTIEG--G--DCSFHFHLvLSEPAICRR--ERRGIF
269
gi 8928214 198 YFK---E-SLIDRVQNVYGNVIENNKFRV--LKKEHDNIKIEiFLAPDNFSK-KSKRHIK
250
130 140 150 160 170 180
....*....|....*....|....*....|....*....|....*....|....*....|
consensus 112 LFINGRPVEDKLLLKAIREVYATYLPRGRYPVFVLNLELPPELVDVNVHPDKKEVRLLKE 171
query 260 LFINHRLVESTSLRKAIETVYAAYLPKNTHPFLYLSLEISPQNVDVNVHPTKHEVHFLHE 319
1B63_A 258 CYVNGRMMRDRLINHAIRQACEDKLGADQQPAFVLYLEIDPHQVDVNVHPAKHEVRFHQS 317
gi 8039787 270 TFVNGRRIFDYGLVQALVLGSEGYFPNGTFPVACLFLTVNSERIDFNIHPAKKEVHLQDY
329
gi 8928214 251 TFVNRRPIDQKDLLEAITNGHSRILSPGNFPICYLFLEINPEYIDFNVHPQKKEVRFYNL
310
....*...
consensus 172 EEILDLIK 179
query 320 ESILERVQ 327
1B63_A 318 RLVHDFIY 325
gi 8039787 330 AHIRHTLS 337
gi 8928214 311 PFLFKLIS 318
Which is shown on another webpage.