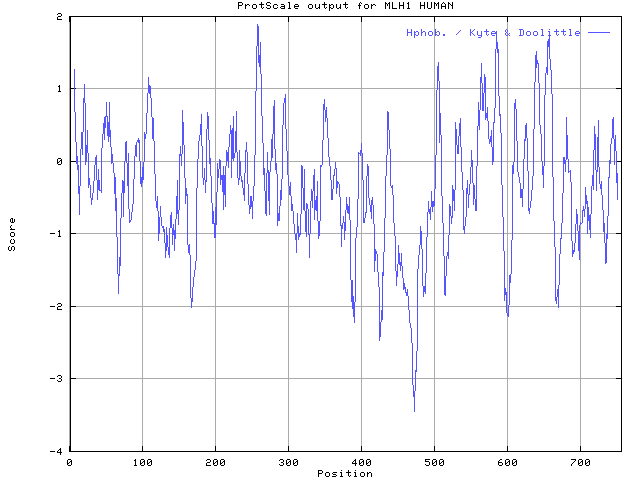
Sequence analysis of MLH1 protein.
- To search protein file in SWISS-PROT Database - http://kr.expasy.org/sprot/
- To use analysis tools in ExPASy - http://kr.expasy.org/srs5/
-Deadline- 11/29/2001
1. Search human MutL protein homolog 1, Mlh1, in SWISS-PROT database. Give its (a) accession number, (b) entry name and (b) release data of last modification.
Start and continue the program, and then key in the "MLH1" in the first line and the "human" in the second line. If change the status of the first line as "GeneName" that given will be better (or if not, there will be five labs indicating masmatch or something else).
SWISS_PROT:MLH1_HUMAN
MLH1_HUMAN
Annotations were last modified in Release 40, October 2001
2. Give its number of amino acids, molecular weight and theoretical pI.
At the bottom of this page, we can find a "Tool" icon follows several links. Go into the "Compute pI/Mw", the third answer is telled. Then submit and find the rest answers.
The sequence MLH1_HUMAN consists of 756 amino acids
Molecular weight: 84600.98
Theoretical pI: 5.51
3. Calculate the total number of negatively charged residues and positively
charged residues.
As above, choose the "ProtParam" after the icon and directly submit.(The answers of 2. can also be released here)
Total number of negatively charged residues (Asp + Glu): 104
Total number of positively charged residues (Arg + Lys): 83
Further more, atomic composition' total atom number' extinction coeffecient
and other information about its property is also shown here.
4. Calculate its hydrophobicity (Kyte & Doolittle scale, window size
11, Relative weight 60%).
As above, choose the "ProScale" after the icon and directly submit...not at all! Change the window size to 11 and reletive weight to 60% first! Then submit twice.

5. Performe the trypsin (higher specificity) cleavage of the protein. (a) How many peptides will you get after cleavage? (b) Give the list of peptides with a mass bigger than 1000 dalton.
As above, choose the "PeptideMass" after the icon.(The enzyme "Trypsin" has been preseted) Change the peptide bigger than 0 and 1000 and the perform twice.
Total nomber of peprides after cleavage: 81 (peptides bigger than 0)
Peptides bigger than 1000:
| mass | position | #MC | peptide sequence |
| 4615.429 |
|
|
AEMLADYFSLEIDEEGNLIG LPLLIDNYVPPLEGLPIFIL R |
| 3115.482 |
|
|
LSEPAPLFDLAMLALDSPES GWTEEDGPK |
| 2862.478 |
|
|
NTHPFLYLSLEISPQNVDVN VHPTK |
| 2804.404 |
|
|
APPKPCAGNQGTQITVEDLF YNIATR |
| 2775.324 |
|
|
EMLHNHSFVGCVNPQWALAQ HQTK |
| 2751.311 |
|
|
QYISEESTLSGQQSEVPGSI PNSWK |
| 2634.410 |
|
|
IINLTSVLSLQEEINEQGHE VLR |
| 2625.413 |
|
|
LDAFLQPLSKPLSSQPQAIV TEDK |
| 2550.328 |
|
|
LSEELFYQILIYDFANFGVL R |
| 2201.124 |
|
|
HFTEDGNILQLANLPDLYK |
| 2140.082 |
|
|
MYFTQTLLPGLAGPSGEMVK |
| 1968.958 |
|
|
QQDEEMLELPAPAEVAAK |
| 1835.002 |
|
|
GEALASISHVAHVTITTK |
| 1833.902 |
|
|
LQSFEDLASISTYGFR |
| 1774.887 |
|
|
HEVHFLHEESILER |
| 1550.901 |
|
|
IAAGEVIQRPANAIK |
| 1532.692 |
|
|
STTSLTSSSTSGSSDK |
| 1525.596 |
|
|
EDSDVEMVEDDSR |
| 1460.684 |
|
|
MNGYISNANYSVK |
| 1410.751 |
|
|
EGLAEYIVEFLK |
| 1408.722 |
|
|
YSVHNAGISFSVK |
| 1338.730 |
|
|
AIETVYAAYLPK |
| 1333.627 |
|
|
LATEVNWDEEK |
| 1327.733 |
|
|
LIQIQDNGTGIR |
| 1300.686 |
|
|
TLPNASTVDNIR |
| 1275.703 |
|
|
CIFLLFINHR |
| 1174.625 |
|
|
WTVEHIVYK |
| 1165.523 |
|
|
EMIENCLDAK |
| 1148.550 |
|
|
ELIEIGCEDK |
| 1119.496 |
|
|
ECAMFYSIR |
| 1092.517 |
|
|
NQSLEGDTTK |
| 1091.504 |
|
|
EDLDIVCER |
| 1003.514 |
|
|
VYAHQMVR |