1. Add MLH1_Human protein to the Biology Workbench. Predict its secondary structure by GOR4.
We first get:
DNA MISMATCH REPAIR PROTEIN MLH1 (MUTL PROTEIN HOMOLOG 1) [Homo sapiens (Human)]
from Protein Tool - Multiple Database search. Then we do the GOR4 -Predict secondary structure of PS.
>MLH1_HUMAN
MSFVAGVIRRLDETVVNRIAAGEVIQRPANAIKEMIENCLDAKSTSIQVI
VKEGGLKLIQIQDNGTGIRKEDLDIVCERFTTSKLQSFEDLASISTYGFR
GEALASISHVAHVTITTKTADGKCAYRASYSDGKLKAPPKPCAGNQGTQI
TVEDLFYNIATRRKALKNPSEEYGKILEVVGRYSVHNAGISFSVKKQGET
VADVRTLPNASTVDNIRSIFGNAVSRELIEIGCEDKTLAFKMNGYISNAN
YSVKKCIFLLFINHRLVESTSLRKAIETVYAAYLPKNTHPFLYLSLEISP
QNVDVNVHPTKHEVHFLHEESILERVQQHIESKLLGSNSSRMYFTQTLLP
GLAGPSGEMVKSTTSLTSSSTSGSSDKVYAHQMVRTDSREQKLDAFLQPL
SKPLSSQPQAIVTEDKTDISSGRARQQDEEMLELPAPAEVAAKNQSLEGD
TTKGTSEMSEKRGPTSSNPRKRHREDSDVEMVEDDSRKEMTAACTPRRRI
INLTSVLSLQEEINEQGHEVLREMLHNHSFVGCVNPQWALAQHQTKLYLL
NTTKLSEELFYQILIYDFANFGVLRLSEPAPLFDLAMLALDSPESGWTEE
DGPKEGLAEYIVEFLKKKAEMLADYFSLEIDEEGNLIGLPLLIDNYVPPL
EGLPIFILRLATEVNWDEEKECFESLSKECAMFYSIRKQYISEESTLSGQ
QSEVPGSIPNSWKWTVEHIVYKALRSHILPPKHFTEDGNILQLANLPDLY
KVFERC
LEGEND:
Alpha Helix = H Beta Sheet
= E Random Coil = C
2. Do a homology searching of MLH1_Human in Genpept Full Release Database.
Import MLH1-like protein of C. elegans, S. cerevisiae, D. melanogaster, R. norvegicus
and M. musculus to your workbench. Run CLUSTALW to get multiple sequence alignment
for these six proteins.
We choose the sequence to do the BLASTP - compare a PS to a PS DB. First we choose the database SwissProt. We can find and inport three sequences:
MLH1_RAT DNA MISMATCH REPAIR PROTEIN MLH1 (MUTL PROTEIN
HOMOLOG 1) [Rattus norvegicus (Rat)]
MLH1_YEAST MUTL PROTEIN HOMOLOG 1 (DNA MISMATCH REPAIR PROTEIN MLH1) [Saccharomyces
cerevisiae (Baker's yeast)]
MLH1_HUMAN DNA MISMATCH REPAIR PROTEIN MLH1 (MUTL PROTEIN HOMOLOG 1) [Homo sapiens
(Human)]
WE then choose another database Genepept and try again, choose the three sequences below:
GENPEPT:7304079 Drosophila melanogaster genomic scaffold
142000013386047 section 5
GENPEPT:3192877 Drosophila melanogaster mutL homolog (Mlh1) gene, complete cds;
GENPEPT:3880333 Caenorhabditis elegans cosmid T28A8, complete sequence;
Then we select all and do the CLUSTALW - multiple sequence alignment. The setting is the remain:
PAIRWISE ALIGNMENT PARAMETERS
Alignment method: AccurateFast (Clustal V)
Accurate method parameters
Weight matrix: PAM seriesBLOSUM seriesGonnet seriesidentity
Gap open penalty: (0.0 - 100.0)
Gap extension penalty: (0.0 - 10.0)
Fast method parameters
K-tuple size: 12
Gap penalty: (1 - 500)
Top diagonals: (1 - 50)
Window size: (1 - 50)
The result (uncolored) is linked to http://life.nthu.edu.tw/~b881611/homeworks/bi06-2.htm.
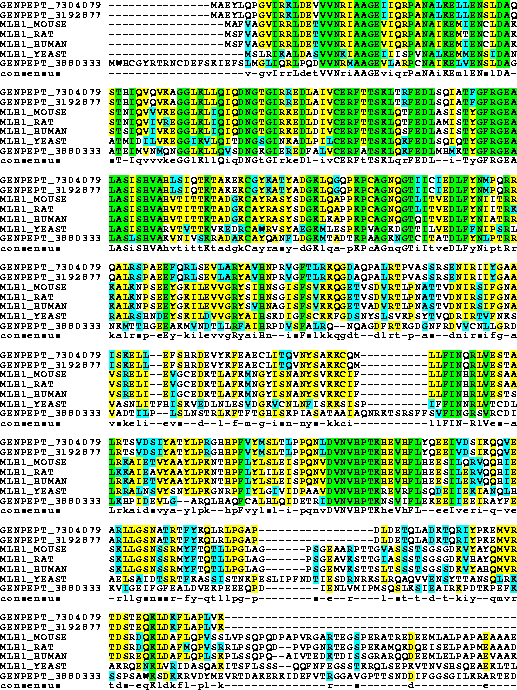
3. Perform BOXSHADE program to get a color-coded plot for the results of
question 2.
After done by the question 2. , we import the alignment. Then pick this alignment and do the BOXSHADE. We keep all the setting they've been given:
Similarity threshhold fraction: 0.5
(0.9 misses many similarities; 0.1 finds false similarities) Lines between Sequence
Blocks:
Show sequence names: yes Residue numbering: None
Character Size: 10 Orientation: Portrait
Show consensus line: yes Consensus Symbols: -LU(different, similar, all-identical)
("L" = lower-case, "U" = upper-case, "B" = blank)
When the ruler is chosen for alignment numbering, boxshade can get stuck and
never finish. Try changing the font size or page orientation when this happens.
--------------------------------------------------------------------------------
Sequence Comparison
Similarity to a Master Sequence?no Number of Master Sequence: 1
Hide Master Sequence? no Show Master Sequence in all-normal Format? no
--------------------------------------------------------------------------------
Shading/Coloring Scheme
Completely Conserved Residues
Background Color: Green Foreground Color: Black Foreground Letter Case: Upper
Identical Residues
Background Color: Yellow Foreground Color: Black Foreground Letter Case: Upper
Similar Residues
Background Color: Cyan Foreground Color: Black Foreground Letter Case: Upper
Different Residues
Background Color: White Foreground Color: Black Foreground Letter Case: Upper
--------------------------------------------------------------------------------
Similiarity Definitions
Boxshade default similarities
Individual Similarities:
D: E F: YW G: A I: LVM L: VMI M: ILV
N: Q R: K T: S V: MIL W: FY Y: WF
Groups:
FYW IVLM RK DE GA TS NQ
We can download it as jpg form to show it:

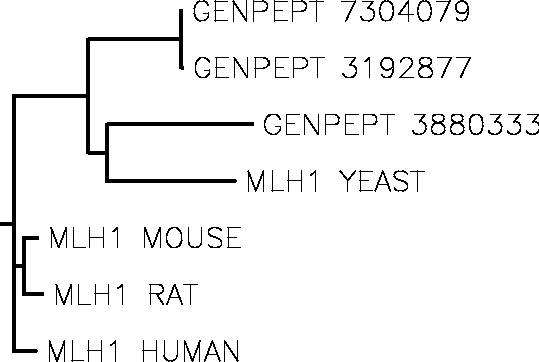
4. Draw rooted phylogenetic tree for these proteins.
In question 2. , we change the Guide tree display to Rooted tree, then we can get the plot below:
Clustal W dendrogram