Protein
Science (2000), 9: 197-200. Cambridge University Press. Printed
in the USA.
Copyright (C) 2000 The Protein Society
Expectations from structural genomics
STEVEN E. BRENNER
1
and MICHAEL LEVITT
1
1 Department of Structural Biology, Stanford
University, Fairchild Building D-109, Stanford, California 94305-5126
The SCOP database organizes proteins according
to their structural and evolutionary relationships.
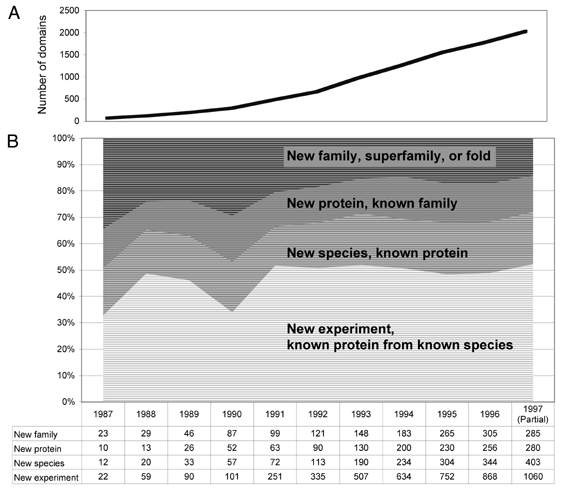
Even as the number of domains studied has grown dramatically, the nature
of the sequences studied has been comparatively constant.
~ 50% - New experiment,
known protein from known species
with some mutations, different conditions, in a larger complex,
or with bound ligands.
~ 20% - New species,
known protein
domains were from a protein for which a structure had been
solved from a different species
~ 14% - New protein,
known family
new proteins for which there was a known structure of a
homolog in the same family.
~ 85% of the new protein domain structures
experimentally determined were in the same SCOP family as a protein already
in the PDB.
Relationships between these proteins could have been recognized by sequence
comparison, and it should have been possible to structurally model
the protein domains by computational methods.
~ 15% - New family or folds
For proteins lacking significant pairwise sequence similarity to those
already in the protein database.

In 1997, fewer than a quarter of such protein
domains had a new fold, compared with about a half
in 1990.
This suggests that the 459 protein folds in
the most recent SCOP incorporate a majority of the frequently occurring
globular structures.
From this trend, it might seem that all of the most
common folds may soon be known.