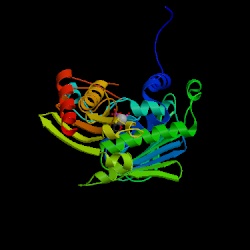
1. Make a GIF picture of the PDB file that you find in assignment 7 by RasMol. Display it in cartoon form and color it by structure. Please make the orientation of the molecule just like the molecule in the picture of assignment 7 - 2.
From the answer of the assingment 7, we take the protein with its ID 1B62 to continue our works. We can key in the 1B62 directly in the site.In the database, we can open its structure in View Structure or download the Rasmol file.
In the Rasmol program, we can use the mouse left button to rotate the molecule and with the Shift key for zooming in or out. We choose the cartoon form in the Desplay line and color it by structure in the Colours. The picture will show the alpha-chains in pink and the beta-sheets in yellow. We can save the picture shown in gif. form or any others in the Export.
Comparing with the figure on the website shown:
2. Residues Glu173 and Arg177 are possible to form an ion pair (salt-bridge).
(a) Draw a zoom-in picture of this pair. Show these two residues in sticks and
color with cpk. The rest of protein in backbone form. (b) Measure the distance
between Glu173A.OE and ARG177A.NH.
In the Rasmol Command Line of the other window, key in the command: select 173,177 to pick the residues(groups) indicated.
Well......, change the whole molecule to the display form Backbone and give it color by structure. Then do the selecting. After change their display, we can easily find them and do operation on them only, like shown below that we can change their color:
If we set the Setting as Picking Distance, the distance between the two atoms we select will be shown on the picture.
3. Show a protein fragment with residues 622 to 699 only. Show them in cartoon
form and color with temperature.
The command is: select 622-699 and restrict 622-699 to display the residues 622-699 or anything selected only.
But right now, we can find that this protein in ID 1B62 has only 355 residues, which means that the protein I found is not the one the teacher expected. But by comparing the two protein, 1B62 and 1BKN, we can find that their structure is alike that we can expect the residues 622 to 699 in 1BKN would have homologous residues in 1B62. By doing the BLASTEQ in Biology WorkBanch, the result would help us to find that the residues in 1B62 that is homologous to the residues 622 to 699 in 1BKN is 222 to 298.
The BLAST data is linked here.
4. Make a stereo picture for only the atoms within 6.0 Angstrom of Phe721. Show
this residue in green color, others in cpk and all the residues in wireframe
0.1 format.
As the method provided in the answer 3. , we can find that the homologous residue in protein 1B62 to the residue Phe721 in the protein 1BKN is Phe320.
The command is: restrict within (6.6,320). I put on the ADP and the Mg atoms for labeling their relative position.