5.7 EVA Dialogs
5.7.1 Add New EVA Column Dialog
Function:
To define the parameters for an EVA column.
License Requirements:
This feature requires a QSAR license ("QSAR").
Menubar:
Dialog Description:
Figure 42 Add New EVA Column dialog
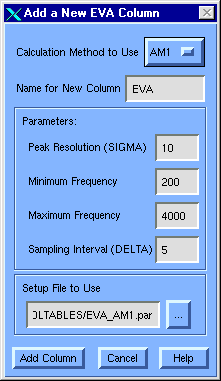
Calculation Method to Use [option menu]
Select the MOPAC Hamiltonian to use for calculating normal modes for each molecule. AM1 and PM3 are the current options.
Name for New Column [field]
Enter the name to give the new column. With the table name, this will be used to create a name for the new directory created to hold setup and results files for each MOPAC calculation job.
Peak Resolution (SIGMA) [field]
Gaussian peak half-width at half-height to use in calculating the EVA profile contribution from each normal mode (see EVA Theory), which corresponds to the spectral resolution in that profile. The default is 10 cm-1.
Minimum Frequency [field]
Frequency at which to begin calculating the EVA profile. The default value is 200 cm-1. Any non-negative value below 4000 can be used, but modes below 200 cm-1 tend to be too dependent on conformation to be generally useful.
Maximum Frequency [field]
Maximum frequency to consider.The default of 4000 cm-1 is the maximum allowed.
Sampling Interval (DELTA) [field]
Each profile is converted into a vector descriptor by sampling at fixed intervals between the Minimum and the Maximum frequency specified. The default is 5 cm-1. This variable must be a positive integer value and should always be less than or equal to 2 , but
, but  /2 is generally optimal.
/2 is generally optimal.
Setup File to Use [field]
Points to a MOPAC setup file to use. For AM1 fields, the default is $TA_MOLTABLES/EVA_AM1.par.
Specify File Names [check box]
If this box is checked, you will be prompted to enter a MOPAC job file name to use for each selected row. Otherwise, SYBYL will generate names based on row numbers.
Remarks:
How long the MOPAC job for each cell in an EVA column will take to run will vary with the dataset in hand and with the current machine configuration (see the Quantum Manual). Column evaluation for structures without 3D coordinates will almost always fail. Moreover, evaluation will generally be much faster and more likely to be successful when the initial structures have been minimized (e.g., Compute >>> Minimize... or MAXIMIN with charges taken into consideration) before trying to create an EVA column.
You can get a feel for how long evaluating the column for the entire table will take by highlighting the first one or two rows and the first empty column, then pressing MSS: AutoFill and working through the Add New EVA Column dialog as described below. This will create the EVA column and evaluate the selected rows. To evaluate the rest of the column, simply deselect (or select) all rows while keeping the EVA column highlighted and press MSS: AutoFill again. It will generally be more convenient, however, to simply let evaluations run overnight.
You can interrupt EVA column evaluation at any time. Doing so will abort evaluation when the job currently being processed completes, but no data from cells which have already been evaluated will be lost.
Each EVA column has an associated Directory and Parameter file; you can see what they are for any given column using MSS: Info >>> Columns. When a cell in an EVA column is evaluated, SYBYL looks in that column's directory for a MOPAC Results file from which to get that molecule's normal modes; you can access the name of that file using MSS: Info >>> Cells. If no such Results file is found, SYBYL launches a MOPAC job using the Parameter file associated with the EVA column.
If initial evaluation fails (typically because the job timed out or the starting structure was too far from equilibrium), you can change the starting structure and force re-evaluation by deleting existing results files. Alternatively, you can run external MOPAC jobs, copy the requisite Results files into the Directory associated with the EVA column, then re-evaluate the cell in question (e.g., using MSS: AutoFill).
Additional Information:


Copyright © 1999, Tripos Inc. All rights
reserved.













 , but
, but  /2 is generally optimal.
/2 is generally optimal.
